

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MVPHLLLLCLLPLVRATEPHEGRADEQSAEAALAVPNASHFFSWNNYTFSDWQNFVGRRRYGAESQNPTVKALLIVAYSFIIVFSLFGNVLVCHVIFKNQRMHSATSLFIVNLAVADIMITLLNTPFTLVRFVNSTWIFGKGMCHVSRFAQYCSLHVSALTLTAIAVDRHQVIMHPLKPRISITKGVIYIAVIWTMATFFSLPHAICQKLFTFKYSEDIVRSLCLPDFPEPADLFWKYLDLATFILLYILPLLIISVAYARVAKKLWLCNMIGDVTTEQYFALRRKKKKTIKMLMLVVVLFALCWFPLNCYVLLLSSKVIRTNNALYFAFHWFAMSSTCYNPFIYCWLNENFRIELKALLSMCQRPPKPQEDRPPSPVPSFRVAWTEKNDGQRAPLANNLLPTSQLQSGKTDLSSVEPIVTMS | |
| CCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHSSSSHHHHHHHHHHHHHHHHHSSSSSCCCCCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 912678876652214899988888888877767889876678888887554567766555676667837999999999999999988898860653007888859999999999999999999899999999818677857999999999999999999999999998589976569765345366625645999999999999998123773015887627997048964669999999999999999999999999999999993167988755267889888530699999999999999989999999999936754479999999999999998999999975999999999998630799853356888877775554356888876777788478777788996765667763589 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MVPHLLLLCLLPLVRATEPHEGRADEQSAEAALAVPNASHFFSWNNYTFSDWQNFVGRRRYGAESQNPTVKALLIVAYSFIIVFSLFGNVLVCHVIFKNQRMHSATSLFIVNLAVADIMITLLNTPFTLVRFVNSTWIFGKGMCHVSRFAQYCSLHVSALTLTAIAVDRHQVIMHPLKPRISITKGVIYIAVIWTMATFFSLPHAICQKLFTFKYSEDIVRSLCLPDFPEPADLFWKYLDLATFILLYILPLLIISVAYARVAKKLWLCNMIGDVTTEQYFALRRKKKKTIKMLMLVVVLFALCWFPLNCYVLLLSSKVIRTNNALYFAFHWFAMSSTCYNPFIYCWLNENFRIELKALLSMCQRPPKPQEDRPPSPVPSFRVAWTEKNDGQRAPLANNLLPTSQLQSGKTDLSSVEPIVTMS | |
| 733100000000003124344333444324321211211131223122233132323444243332210000000210020010023132000000000340200000001000201100010001000000003200101000000000000000000000000000001000000122213310000000011101000100000010131413743210000010134342010000001002112310110020001000102333332544454344334321000000000000000002020000000002323012000000000001000000000000143004002300200213444465344343433334344444344443444413444344453433333233438 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHSSSSHHHHHHHHHHHHHHHHHSSSSSCCCCCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MVPHLLLLCLLPLVRATEPHEGRADEQSAEAALAVPNASHFFSWNNYTFSDWQNFVGRRRYGAESQNPTVKALLIVAYSFIIVFSLFGNVLVCHVIFKNQRMHSATSLFIVNLAVADIMITLLNTPFTLVRFVNSTWIFGKGMCHVSRFAQYCSLHVSALTLTAIAVDRHQVIMHPLKPRISITKGVIYIAVIWTMATFFSLPHAICQKLFTFKYSEDIVRSLCLPDFPEPADLFWKYLDLATFILLYILPLLIISVAYARVAKKLWLCNMIGDVTTEQYFALRRKKKKTIKMLMLVVVLFALCWFPLNCYVLLLSSKVIRTNNALYFAFHWFAMSSTCYNPFIYCWLNENFRIELKALLSMCQRPPKPQEDRPPSPVPSFRVAWTEKNDGQRAPLANNLLPTSQLQSGKTDLSSVEPIVTMS | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.24 | 0.21 | 0.70 | 2.80 | Download | ----------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.24 | 0.23 | 0.70 | 3.79 | Download | ---------------------------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP------------------------------------------------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.25 | 0.21 | 0.70 | 3.72 | Download | ----------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.18 | 0.21 | 0.85 | 1.51 | Download | ------------------------------------DLRDNETWWYNPSIIVH----PHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTL----EGVLCNCSFDYISRDST-TRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMARKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETED-------DKDAET---EIPAGESSDAAPSADAAQMKE---------- | |||||||||||||||||||
| 5 | 4djh | 0.26 | 0.23 | 0.69 | 1.16 | Download | ------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNTKPNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA--LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.21 | 0.23 | 0.82 | 3.24 | Download | -PEMKDFRHGFDILVGQIDDALKLANEG-----KVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.20 | 0.19 | 0.67 | 1.72 | Download | ----------------------------------------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRKMAGMMIAAAWVLSFILWAPAILFWQFIVG--VRTVEDGECYIQFFSN-----AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGGAAWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.28 | 0.19 | 0.65 | 4.90 | Download | ------------------------------------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLLT-LPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNV-----AIWALASVVGVPVAIM---GSAQVEDEEIE--CLVEIPTPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS-----REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR------------------------------------------------------------------ | |||||||||||||||||||
| 9 | 4n6hA | 0.24 | 0.21 | 0.70 | 3.04 | Download | ----------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.27 | 0.21 | 0.69 | 2.97 | Download | -------------------------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQ-----GSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS-----KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
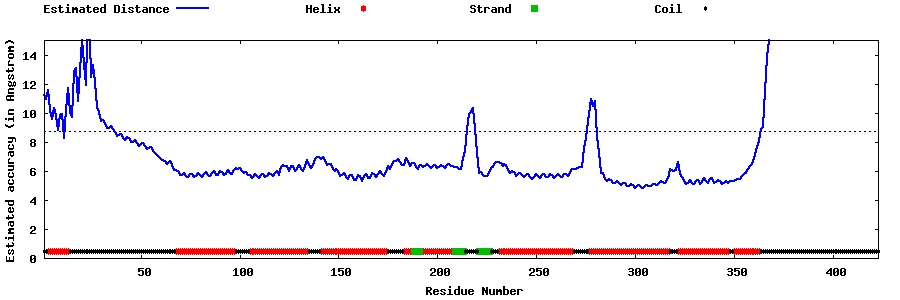
| Generated 3D models | Estimated local accuracy of models | ||
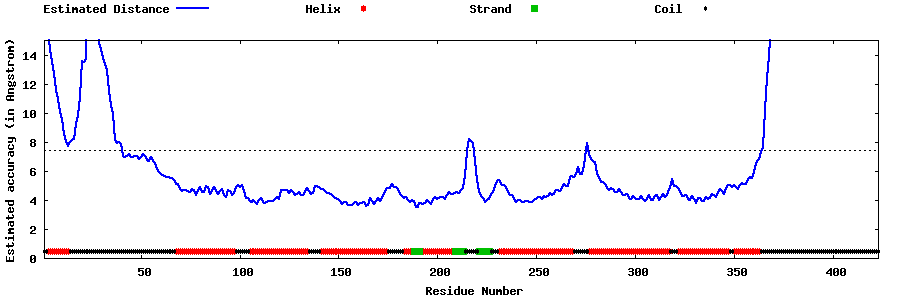 | |||
|
|
|||
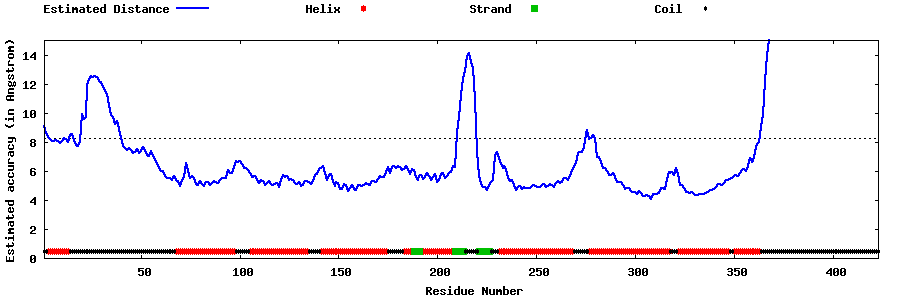 | |||
|
| |||
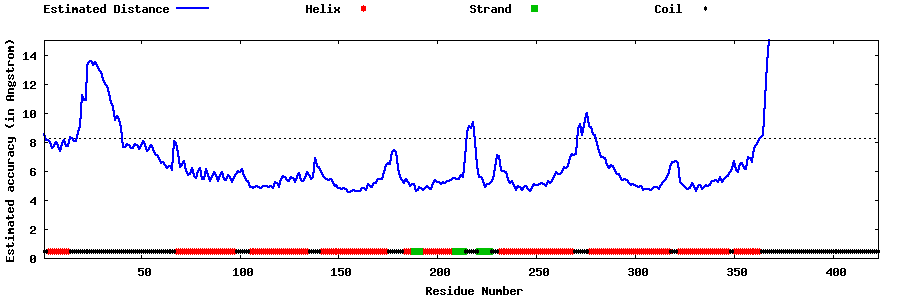 | |||
|
| |||
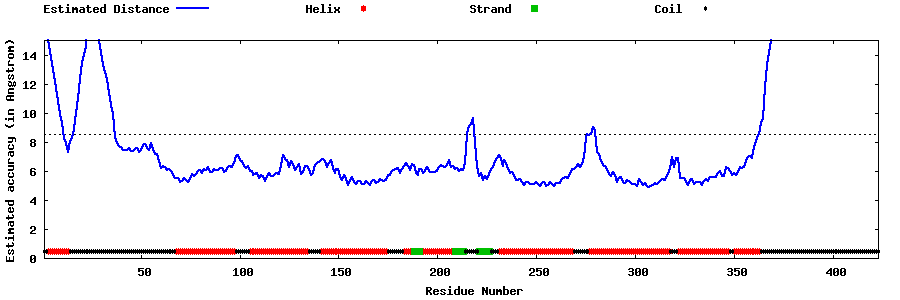 | |||
|
| |||
 | |||
|
| |||