

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MERKFMSLQPSISVSEMEPNGTFSNNNSRNCTIENFKREFFPIVYLIIFFWGVLGNGLSIYVFLQPYKKSTSVNVFMLNLAISDLLFISTLPFRADYYLRGSNWIFGDLACRIMSYSLYVNMYSSIYFLTVLSVVRFLAMVHPFRLLHVTSIRSAWILCGIIWILIMASSIMLLDSGSEQNGSVTSCLELNLYKIAKLQTMNYIALVVGCLLPFFTLSICYLLIIRVLLKVEVPESGLRVSHRKALTTIIITLIIFFLCFLPYHTLRTVHLTTWKVGLCKDRLHKALVITLALAAANACFNPLLYYFAGENFKDRLKSALRKGHPQKAKTKCVFPVSVWLRKETRV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9966578899987666788888788887878737667800899999999999998999778442257878599999999999999999978999999837999887866799999999999999999999999985899970412443556023544679999999999988746888953998899857985258999999999999999999999999999999983578886532000442477743444355797658999999999923666247999999999999999999999999992489999999999720367887656478998778756859 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MERKFMSLQPSISVSEMEPNGTFSNNNSRNCTIENFKREFFPIVYLIIFFWGVLGNGLSIYVFLQPYKKSTSVNVFMLNLAISDLLFISTLPFRADYYLRGSNWIFGDLACRIMSYSLYVNMYSSIYFLTVLSVVRFLAMVHPFRLLHVTSIRSAWILCGIIWILIMASSIMLLDSGSEQNGSVTSCLELNLYKIAKLQTMNYIALVVGCLLPFFTLSICYLLIIRVLLKVEVPESGLRVSHRKALTTIIITLIIFFLCFLPYHTLRTVHLTTWKVGLCKDRLHKALVITLALAAANACFNPLLYYFAGENFKDRLKSALRKGHPQKAKTKCVFPVSVWLRKETRV | |
| 8456245244433444243333343442441336402200002201200230332321000000122343310100000000000000000001000003444210010010000011131111000000000310010001002023223331000000000100110000000023346422100000234731211001011112012212210020011001101214356464444210000000000000100123101010010012334413410100010010001300031020000004401520140035334444444243443433454246 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCC MERKFMSLQPSISVSEMEPNGTFSNNNSRNCTIENFKREFFPIVYLIIFFWGVLGNGLSIYVFLQPYKKSTSVNVFMLNLAISDLLFISTLPFRADYYLRGSNWIFGDLACRIMSYSLYVNMYSSIYFLTVLSVVRFLAMVHPFRLLHVTSIRSAWILCGIIWILIMASSIMLLDSGSEQNGSVTSCLELNLYKIAKLQTMNYIALVVGCLLPFFTLSICYLLIIRVLLKVEVPESGLRVSHRKALTTIIITLIIFFLCFLPYHTLRTVHLTTWKVGLCKDRLHKALVITLALAAANACFNPLLYYFAGENFKDRLKSALRKGHPQKAKTKCVFPVSVWLRKETRV | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.25 | 0.25 | 0.86 | 3.36 | Download | -------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPDGAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.24 | 0.24 | 0.84 | 4.06 | Download | ---------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSKEGLHYTCSSHFPQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKEEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.25 | 0.25 | 0.86 | 3.86 | Download | -------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPDGAVVCMLFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.19 | 0.83 | 1.53 | Download | -------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKE----RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN--QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------- | |||||||||||||||||||
| 5 | 4djh | 0.27 | 0.26 | 0.81 | 1.16 | Download | ---------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKDVDVIECSLQFPDDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS--A-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.25 | 0.25 | 0.86 | 3.41 | Download | ---------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRGAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| 7 | 3uon | 0.18 | 0.20 | 0.79 | 1.66 | Download | -------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFMNLYTL-YTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFITVEDGECYIQFFS----NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIGYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP-C----IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.25 | 0.23 | 0.82 | 4.75 | Download | --------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFRSQKEGLHYTCSSHFPYSQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK--------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------- | |||||||||||||||||||
| 9 | 5o9hA | 0.26 | 0.23 | 0.84 | 3.48 | Download | -------------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFE-AKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREPPKVLCGVDHD--KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSAR------ETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR--------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.24 | 0.25 | 0.93 | 3.97 | Download | NEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVAVTRPRDGAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
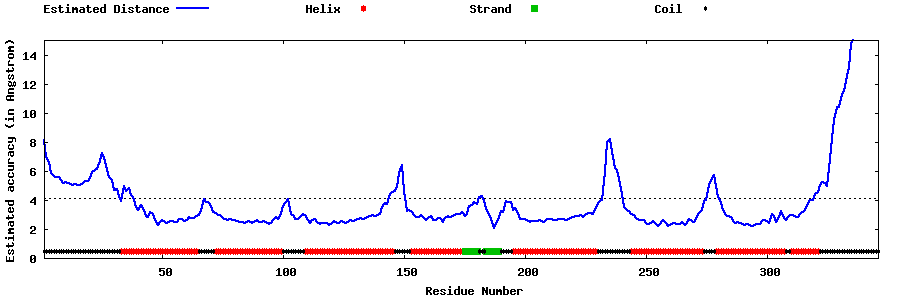 | |||
|
|
|||
 | |||
|
| |||
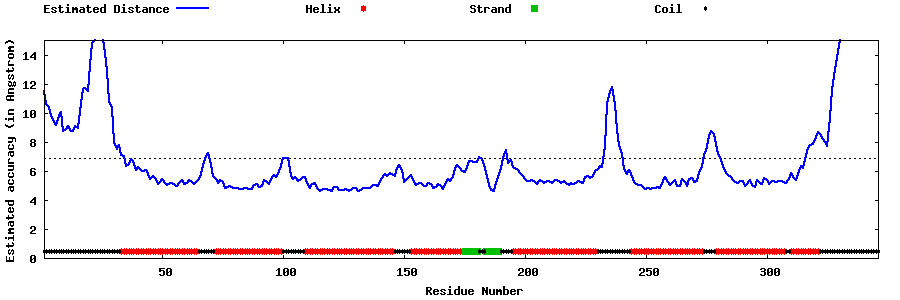 | |||
|
| |||
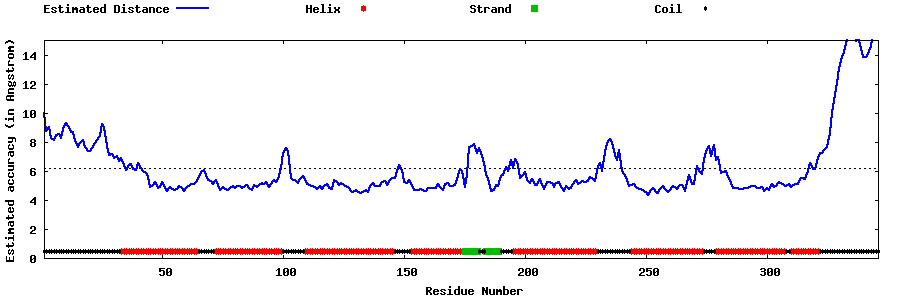 | |||
|
| |||
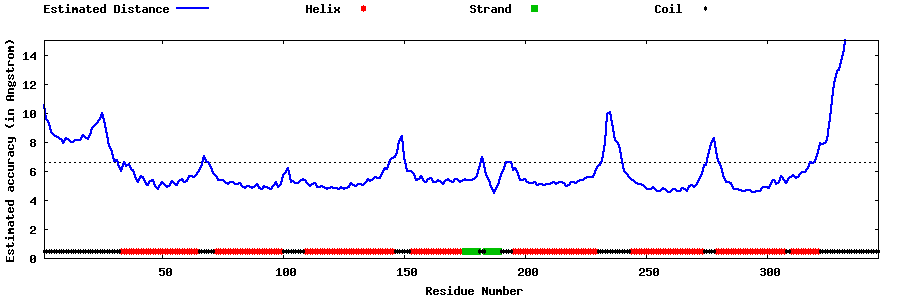 | |||
|
| |||