

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MANASEPGGSGGGEAAALGLKLATLSLLLCVSLAGNVLFALLIVRERSLHRAPYYLLLDLCLADGLRALACLPAVMLAARRAAAAAGAPPGALGCKLLAFLAALFCFHAAFLLLGVGVTRYLAIAHHRFYAERLAGWPCAAMLVCAAWALALAAAFPPVLDGGGDDEDAPCALEQRPDGAPGALGFLLLLAVVVGATHLVYLRLLFFIHDRRKMRPARLVPAVSHDWTFHGPGATGQAAANWTAGFGRGPTPPALVGIRPAGPGRGARRLLVLEEFKTEKRLCKMFYAVTLLFLLLWGPYVVASYLRVLVRPGAVPQAYLTASVWLTFAQAGINPVVCFLFNRELRDCFRAQFPCCQSPRTTQATHPCDLKGIGL | |
| CCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSCCCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHSSSSSSSSSHHHHHCCHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCC | |
| 997899999988761799999999999999999999984220461476653758999999999999999986599999987400575327603444379989999999999999999998998106142454344488999999999999999999999804875787726999637874245136678999999999999999999999998773233312232222346531112212344211022455443211234543321000233344444210167777550689855599999999998066678579999999999999999899998078999999999837779999887789988778999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MANASEPGGSGGGEAAALGLKLATLSLLLCVSLAGNVLFALLIVRERSLHRAPYYLLLDLCLADGLRALACLPAVMLAARRAAAAAGAPPGALGCKLLAFLAALFCFHAAFLLLGVGVTRYLAIAHHRFYAERLAGWPCAAMLVCAAWALALAAAFPPVLDGGGDDEDAPCALEQRPDGAPGALGFLLLLAVVVGATHLVYLRLLFFIHDRRKMRPARLVPAVSHDWTFHGPGATGQAAANWTAGFGRGPTPPALVGIRPAGPGRGARRLLVLEEFKTEKRLCKMFYAVTLLFLLLWGPYVVASYLRVLVRPGAVPQAYLTASVWLTFAQAGINPVVCFLFNRELRDCFRAQFPCCQSPRTTQATHPCDLKGIGL | |
| 610213344343231010001110112022103323200000001133013000000000010000000000100000002221233021120000000000000000001020010031100002003132321420000000000000000001000000113344230203232210000001322223333213202320000010212232343233233334334443344443434443444334443343444343433334333443444320000000001101100200000000100034230130000000110120001000000000440040024002012243444643426266434 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSCCCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHSSSSSSSSSHHHHHCCHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCC MANASEPGGSGGGEAAALGLKLATLSLLLCVSLAGNVLFALLIVRERSLHRAPYYLLLDLCLADGLRALACLPAVMLAARRAAAAAGAPPGALGCKLLAFLAALFCFHAAFLLLGVGVTRYLAIAHHRFYAERLAGWPCAAMLVCAAWALALAAAFPPVLDGGGDDEDAPCALEQRPDGAPGALGFLLLLAVVVGATHLVYLRLLFFIHDRRKMRPARLVPAVSHDWTFHGPGATGQAAANWTAGFGRGPTPPALVGIRPAGPGRGARRLLVLEEFKTEKRLCKMFYAVTLLFLLLWGPYVVASYLRVLVRPGAVPQAYLTASVWLTFAQAGINPVVCFLFNRELRDCFRAQFPCCQSPRTTQATHPCDLKGIGL | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.17 | 0.18 | 0.77 | 2.42 | Download | -ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVF-----HRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTR-PKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCS--DIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKA-------------------------------------------------------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------------------- | |||||||||||||||||||
| 2 | 4z34A | 0.14 | 0.21 | 0.91 | 4.18 | Download | --NRSGKHLATEWN-TVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNT----RRLTVS-TWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRM--SNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKAMKDQAAAEQLKTTRNAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVLAY-EKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQIL---------------------- | |||||||||||||||||||
| 3 | 5uenA | 0.16 | 0.20 | 0.93 | 3.11 | Download | ------------SISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIG----PQTYF--HTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTP-RRAAVAIAGCWILSFVVGLTPMFGWNGSMGEPVIKCEFEKVISMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNNVKDALTKMRAAALDAPEMKDFRANEGKVKEAQAAAEQLKTTRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCHKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRCQPLEVLF--------- | |||||||||||||||||||
| 4 | 4ib4 | 0.21 | 0.24 | 0.91 | 1.55 | Download | -----------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFE---AMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFI-KITVVWLISIGIAIPVPKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQMRAALDAQKKHNEGKVKEAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----------------- | |||||||||||||||||||
| 5 | 3uon | 0.18 | 0.24 | 0.89 | 1.20 | Download | --------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVI----GYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTT-KMAGMMIAAAWVLSFILWAPAILFWQFIVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKDTEGTKSPSIGRNTNGVITKNAKLLQQKRWDEAVNLAKSTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPC-IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--------------------- | |||||||||||||||||||
| 6 | 5uenA | 0.15 | 0.20 | 0.92 | 2.95 | Download | -------------ISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIG----PQTYFH--TCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVT-PRRAAVAIAGCWILSFVVGLTPMFGWNNLSSMGEPVIKCEFEKYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDNVKDALTKGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCHKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRCQPLEVLF--------- | |||||||||||||||||||
| 7 | 4ib4 | 0.23 | 0.24 | 0.89 | 1.77 | Download | -----------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMF---EAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNS-RATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERF--GDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDLKIEKADNAAQVAALDKKGFDINEGKVKEAQAAAEQTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNY------------------ | |||||||||||||||||||
| 8 | 4bvnA | 0.21 | 0.22 | 0.75 | 4.02 | Download | ------------LSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRG----TWLWGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLM-TRARAKVIICTVWAISALVSFLPIMMHWWRDEDDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRKRKTS------------------------------------------------------RVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNR-DLVPKWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLL-A-------------------- | |||||||||||||||||||
| 9 | 4ib4A | 0.22 | 0.24 | 0.91 | 2.50 | Download | -----------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMF---EAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNS-RATAFIKITVVWLISIGIAIPVPIKGIETNPNNTCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAGKVKEAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----------------- | |||||||||||||||||||
| 10 | 4nc3A | 0.22 | 0.24 | 0.91 | 2.48 | Download | -----------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIM---FEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNS-RATAFIKITVVWLISIGIAIPVPIKGIDNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRNEGKVKEAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
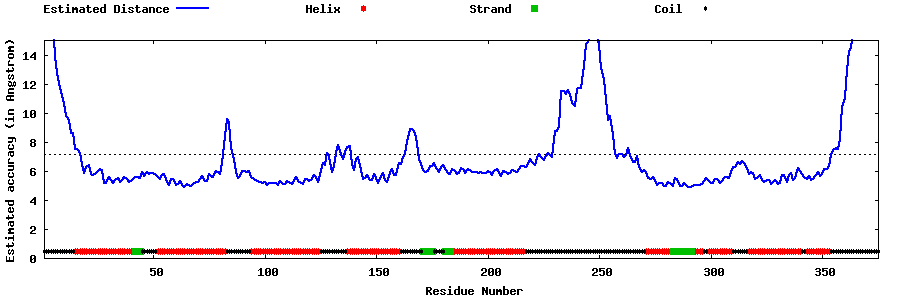
| Generated 3D models | Estimated local accuracy of models | ||
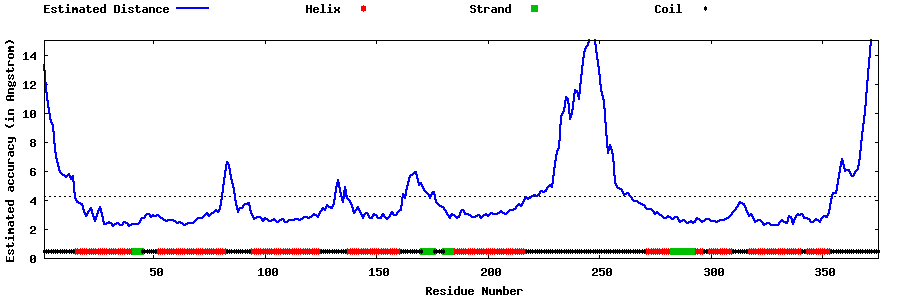 | |||
|
|
|||
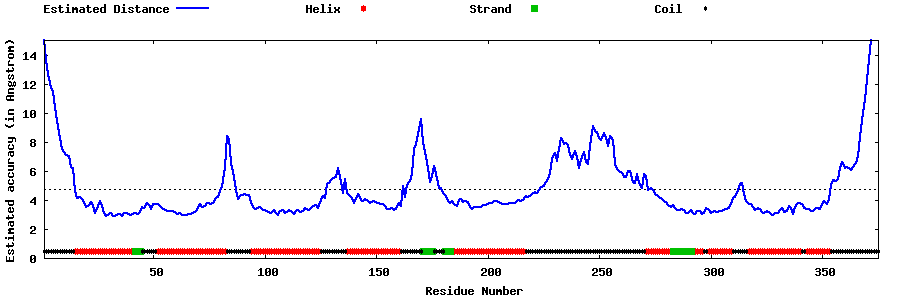 | |||
|
| |||
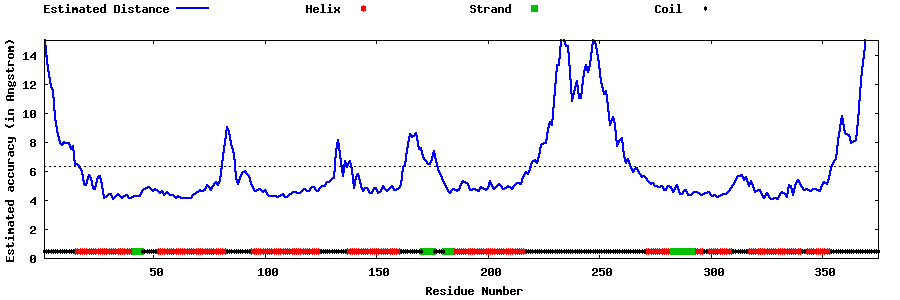 | |||
|
| |||
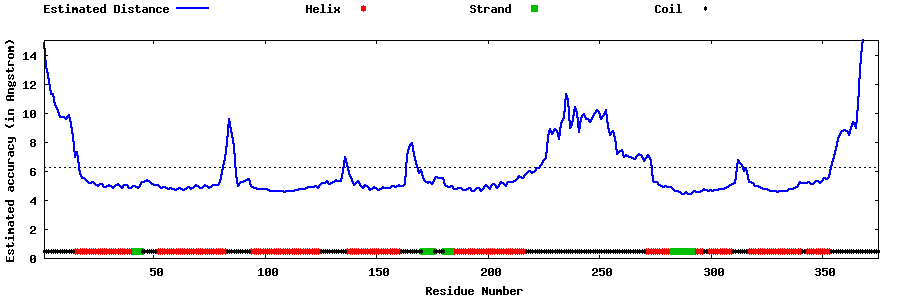 | |||
|
| |||
 | |||
|
| |||