

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MAPSHRASQVGFCPTPERPLWRLPPTCRPRRMSVCYRPPGNETLLSWKTSRATGTAFLLLAALLGLPGNGFVVWSLAGWRPARGRPLAATLVLHLALADGAVLLLTPLFVAFLTRQAWPLGQAGCKAVYYVCALSMYASVLLTGLLSLQRCLAVTRPFLAPRLRSPALARRLLLAVWLAALLLAVPAAVYRHLWRDRVCQLCHPSPVHAAAHLSLETLTAFVLPFGLMLGCYSVTLARLRGARWGSGRHGARVGRLVSAIVLAFGLLWAPYHAVNLLQAVAALAPPEGALAKLGGAGQAARAGTTALAFFSSSVNPVLYVFTAGDLLPRAGPRFLTRLFEGSGEARGGGRSREGTMELRTTPQLKVVGQGRGNGDPGGGMEKDGPEWDL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSHHSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 97200356667688999976679999999888767799987655442079999999999999999876887344100134777777999999999999999999879999999638987862789899999999999999999999997378876130024454557788888999999999995998850477168479967974699999999999999999999999999999999605456654344167855556799999872899999999998743577405789999999999999999999999999999658999999999999986156766678888778762315688765535677899999887889998879 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MAPSHRASQVGFCPTPERPLWRLPPTCRPRRMSVCYRPPGNETLLSWKTSRATGTAFLLLAALLGLPGNGFVVWSLAGWRPARGRPLAATLVLHLALADGAVLLLTPLFVAFLTRQAWPLGQAGCKAVYYVCALSMYASVLLTGLLSLQRCLAVTRPFLAPRLRSPALARRLLLAVWLAALLLAVPAAVYRHLWRDRVCQLCHPSPVHAAAHLSLETLTAFVLPFGLMLGCYSVTLARLRGARWGSGRHGARVGRLVSAIVLAFGLLWAPYHAVNLLQAVAALAPPEGALAKLGGAGQAARAGTTALAFFSSSVNPVLYVFTAGDLLPRAGPRFLTRLFEGSGEARGGGRSREGTMELRTTPQLKVVGQGRGNGDPGGGMEKDGPEWDL | |
| 73663424614403244330242323232442323231443633132400100002100200220232121000000001343311000000000010100000000000000024331012001100000010011000000000010000000000202321120001000000100000001000000203645201000134203100000102102321120002000000110131344664431000000000000010031111000001002303224333131220020010100000030002100000000450043004100220243244634444445334444435435434454444444443547466264 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSHHSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MAPSHRASQVGFCPTPERPLWRLPPTCRPRRMSVCYRPPGNETLLSWKTSRATGTAFLLLAALLGLPGNGFVVWSLAGWRPARGRPLAATLVLHLALADGAVLLLTPLFVAFLTRQAWPLGQAGCKAVYYVCALSMYASVLLTGLLSLQRCLAVTRPFLAPRLRSPALARRLLLAVWLAALLLAVPAAVYRHLWRDRVCQLCHPSPVHAAAHLSLETLTAFVLPFGLMLGCYSVTLARLRGARWGSGRHGARVGRLVSAIVLAFGLLWAPYHAVNLLQAVAALAPPEGALAKLGGAGQAARAGTTALAFFSSSVNPVLYVFTAGDLLPRAGPRFLTRLFEGSGEARGGGRSREGTMELRTTPQLKVVGQGRGNGDPGGGMEKDGPEWDL | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.26 | 0.25 | 0.84 | 2.64 | Download | IDDALKLANEGKVKEAQAAAEQLKTT--RNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYT--KMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-------DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP---------------------------------------------------- | |||||||||||||||||||
| 2 | 5ungA | 0.23 | 0.22 | 0.76 | 3.70 | Download | -----------------------------------YLGSGSCSQKPSDKHLDAIPILYYIIFVIGFLVNIVVVTLF--CCQKGPKKVSSIYIFNLAVADLLLLATLPLWATYYSYRDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFLSQR-RNPWQASYIVPLVWCMACLSSLPTFYFRDVRTIEYCIMAFPPEKWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSYGKNRRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMGVINS--CEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVF------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.27 | 0.24 | 0.75 | 3.74 | Download | --------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI-------DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.25 | 0.23 | 0.71 | 1.50 | Download | ----------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.25 | 0.23 | 0.71 | 1.16 | Download | ----------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR--YTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-----A--------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.20 | 0.18 | 0.74 | 3.11 | Download | ----------------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYK--RLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA-QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK-KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN--NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------ | |||||||||||||||||||
| 7 | 4djh | 0.25 | 0.23 | 0.70 | 1.71 | Download | ------------------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---A----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC------------------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.21 | 0.17 | 0.74 | 4.28 | Download | ---------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYK--RLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ-WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRE-----KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN--CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------ | |||||||||||||||||||
| 9 | 4mbsA | 0.20 | 0.18 | 0.75 | 3.17 | Download | ---------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYK--RLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA-AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK-EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN--CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.23 | 0.19 | 0.73 | 3.40 | Download | -----------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIV--RYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET--------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF----------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
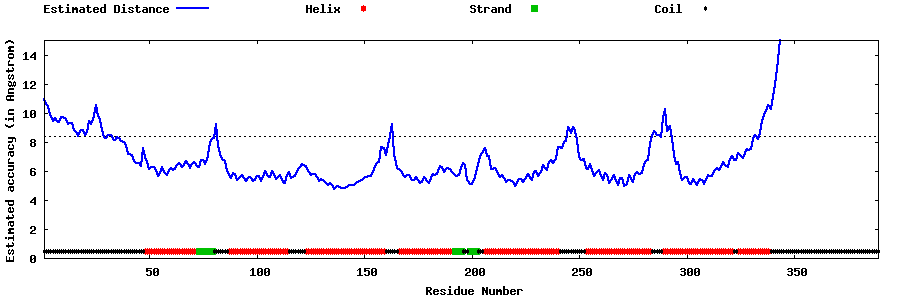
| Generated 3D models | Estimated local accuracy of models | ||
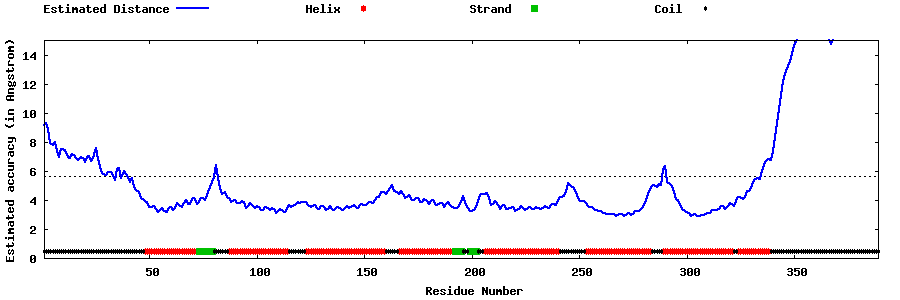 | |||
|
|
|||
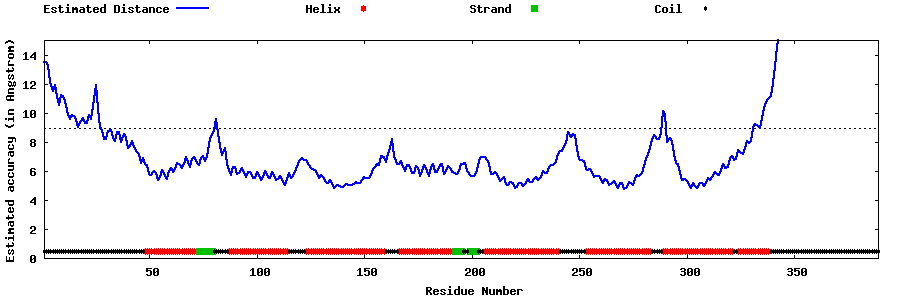 | |||
|
| |||
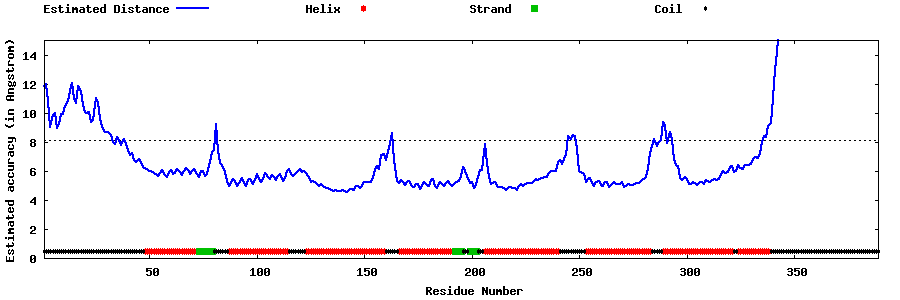 | |||
|
| |||
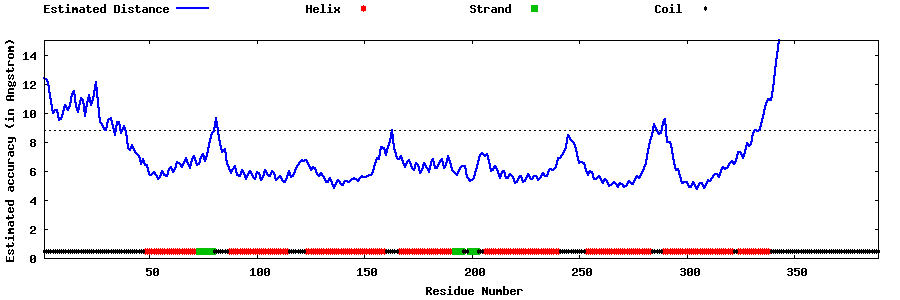 | |||
|
| |||
 | |||
|
| |||