

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MVIMGQCYYNETIGFFYNNSGKELSSHWRPKDVVVVALGLTVSVLVLLTNLLVIAAIASNRRFHQPIYYLLGNLAAADLFAGVAYLFLMFHTGPRTARLSLEGWFLRQGLLDTSLTASVATLLAIAVERHRSVMAVQLHSRLPRGRVVMLIVGVWVAALGLGLLPAHSWHCLCALDRCSRMAPLLSRSYLAVWALSSLLVFLLMVAVYTRIFFYVRRRVQRMAEHVSCHPRYRETTLSLVKTVVIILGAFVVCWTPGQVVLLLDGLGCESCNVLAVEKYFLLLAEANSLVNAAVYSCRDAEMRRTFRRLLCCACLRQSTRESVHYTSSAQGGASTRIMLPENGHPLMDSTL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCCCSSHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 976567657787655678988747788788999999999999999999888721014646878858999999999999999999999999988626155778405197999999999999999999974201677776666419878997879999999999999876350158986442035555526799866999999999999999999999998777515441076767788899999999999999989999999999967887763899999999999999885999996599999999999578889998767677787778998878787999998777899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MVIMGQCYYNETIGFFYNNSGKELSSHWRPKDVVVVALGLTVSVLVLLTNLLVIAAIASNRRFHQPIYYLLGNLAAADLFAGVAYLFLMFHTGPRTARLSLEGWFLRQGLLDTSLTASVATLLAIAVERHRSVMAVQLHSRLPRGRVVMLIVGVWVAALGLGLLPAHSWHCLCALDRCSRMAPLLSRSYLAVWALSSLLVFLLMVAVYTRIFFYVRRRVQRMAEHVSCHPRYRETTLSLVKTVVIILGAFVVCWTPGQVVLLLDGLGCESCNVLAVEKYFLLLAEANSLVNAAVYSCRDAEMRRTFRRLLCCACLRQSTRESVHYTSSAQGGASTRIMLPENGHPLMDSTL | |
| 643134112031232120312343343320110000001110021033133000000010330000000000000100021131123000003232121021000010000000000013000000100000001022344123310000001011212220210000001234243001010123210000001313311210020011001100320442463554444334331000000000010021031111000000001242200200000001001200131100000115400300110000121246465444334435444334434464534346457 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCCCSSHHHHHHHHHHHHHHHHHHHHHHHCCCSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MVIMGQCYYNETIGFFYNNSGKELSSHWRPKDVVVVALGLTVSVLVLLTNLLVIAAIASNRRFHQPIYYLLGNLAAADLFAGVAYLFLMFHTGPRTARLSLEGWFLRQGLLDTSLTASVATLLAIAVERHRSVMAVQLHSRLPRGRVVMLIVGVWVAALGLGLLPAHSWHCLCALDRCSRMAPLLSRSYLAVWALSSLLVFLLMVAVYTRIFFYVRRRVQRMAEHVSCHPRYRETTLSLVKTVVIILGAFVVCWTPGQVVLLLDGLGCESCNVLAVEKYFLLLAEANSLVNAAVYSCRDAEMRRTFRRLLCCACLRQSTRESVHYTSSAQGGASTRIMLPENGHPLMDSTL | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.25 | 0.24 | 0.82 | 2.86 | Download | ----------------ENFMDIECFMVLNSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAG-------KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------- | |||||||||||||||||||
| 2 | 4z34A | 0.54 | 0.49 | 0.87 | 4.21 | Download | ----PQCFYNESIAFFYNRSGKHLATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWNAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVL-AYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQIL----------------------------------------- | |||||||||||||||||||
| 3 | 5tgzA | 0.24 | 0.25 | 0.82 | 3.29 | Download | ----------GGRGENFMDIECFMVLN-PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDK--TYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA--------PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------- | |||||||||||||||||||
| 4 | 4z34 | 0.54 | 0.49 | 0.87 | 1.66 | Download | --NEPQCFYNESIAFFYNRSGKHLATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTR-MNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWEKAMKKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDV-LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG---------------------------------------- | |||||||||||||||||||
| 5 | 4z34 | 0.54 | 0.49 | 0.88 | 1.18 | Download | --NEPQCFYNESIAFFYNRSGKHLATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETKAMKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDV-LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG---------------------------------------- | |||||||||||||||||||
| 6 | 5tjvA | 0.25 | 0.24 | 0.81 | 3.10 | Download | ------------------MDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRITRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAG-------KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------- | |||||||||||||||||||
| 7 | 4z34 | 0.54 | 0.49 | 0.87 | 1.83 | Download | ----PQCFYNESIAFFYNRSGKHLATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLERKAMKDFQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQC-DVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG---------------------------------------- | |||||||||||||||||||
| 8 | 1u19A | 0.18 | 0.23 | 0.93 | 4.40 | Download | ---MNGTEGPNFYVPFSNKTGVVRSPFEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWQCSCGIDYYTPHETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAA----AQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTTETSQVAPA----------------- | |||||||||||||||||||
| 9 | 5tjvA | 0.25 | 0.24 | 0.82 | 3.18 | Download | ----------------ENFMDECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKA--GKRAMSF-----SDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------- | |||||||||||||||||||
| 10 | 5g53A | 0.25 | 0.23 | 0.79 | 3.59 | Download | -------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS-TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCCGEGQVACLFEVVPMNYMVYFNFFVLVPLLLMLGVYLRIFLAARRQLKQMT---------LQKEVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDCSHPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLENLY-------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
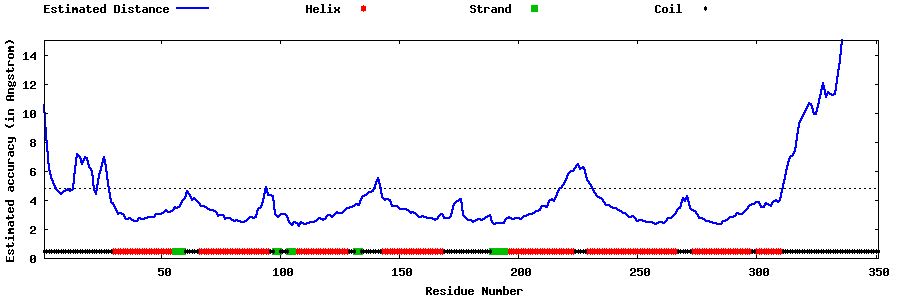 | |||
|
|
|||
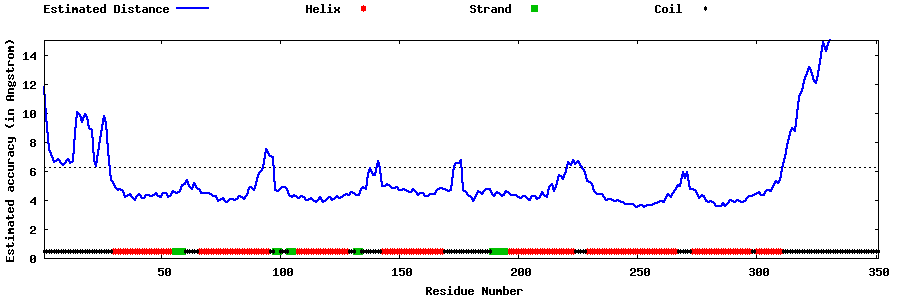 | |||
|
| |||
 | |||
|
| |||
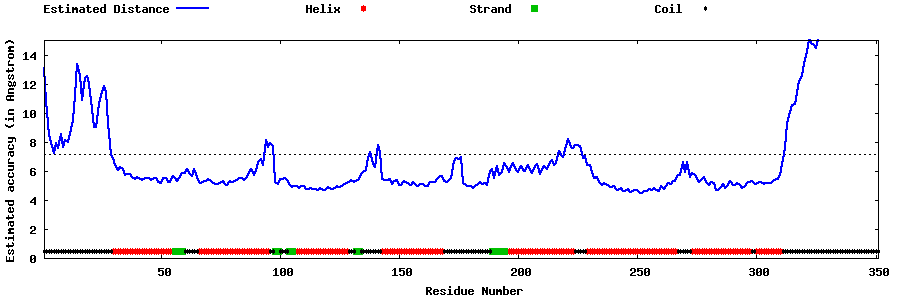 | |||
|
| |||
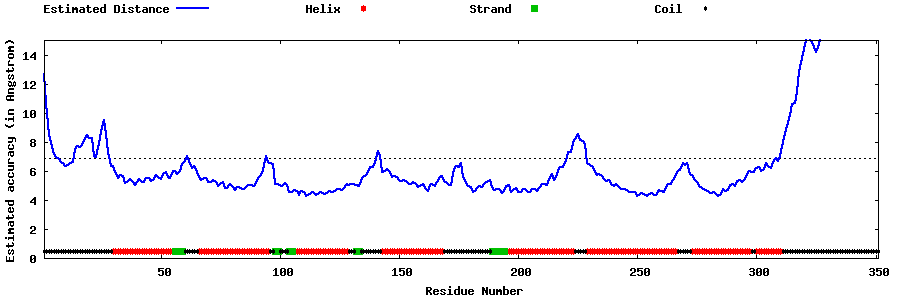 | |||
|
| |||