

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MTPLCLNCSVLPGDLYPGGARNPMACNGSAARGHFDPEDLNLTDEALRLKYLGPQQTELFMPICATYLLIFVVGAVGNGLTCLVILRHKAMRTPTNYYLFSLAVSDLLVLLVGLPLELYEMWHNYPFLLGVGGCYFRTLLFEMVCLASVLNVTALSVERYVAVVHPLQARSMVTRAHVRRVLGAVWGLAMLCSLPNTSLHGIRQLHVPCRGPVPDSAVCMLVRPRALYNMVVQTTALLFFCLPMAIMSVLYLLIGLRLRRERLLLMQEAKGRGSAAARSRYTCRLQQHDRGRRQVTKMLFVLVVVFGICWAPFHADRVMWSVVSQWTDGLHLAFQHVHVISGIFFYLGSAANPVLYSLMSSRFRETFQEALCLGACCHRLRPRHSSHSLSRMTTGSTLCDVGSLGSWVHPLAGNDGPEAQQETDPS | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCCSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 997655478898787999888988897888888888766788764345566788763089999999999999987888876152152599884889999999999999999999999999995898559851200499999999999999999999985852014155650101998999999999999999999999843574332467778886331376863687789727743638999999999999999999544454333333455544455312778888887878999999999999999699999999999823136740159999999999999999999999999679999999999860403678889888888877544478533679998630168789898866689999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MTPLCLNCSVLPGDLYPGGARNPMACNGSAARGHFDPEDLNLTDEALRLKYLGPQQTELFMPICATYLLIFVVGAVGNGLTCLVILRHKAMRTPTNYYLFSLAVSDLLVLLVGLPLELYEMWHNYPFLLGVGGCYFRTLLFEMVCLASVLNVTALSVERYVAVVHPLQARSMVTRAHVRRVLGAVWGLAMLCSLPNTSLHGIRQLHVPCRGPVPDSAVCMLVRPRALYNMVVQTTALLFFCLPMAIMSVLYLLIGLRLRRERLLLMQEAKGRGSAAARSRYTCRLQQHDRGRRQVTKMLFVLVVVFGICWAPFHADRVMWSVVSQWTDGLHLAFQHVHVISGIFFYLGSAANPVLYSLMSSRFRETFQEALCLGACCHRLRPRHSSHSLSRMTTGSTLCDVGSLGSWVHPLAGNDGPEAQQETDPS | |
| 742411000223141323333121221112233313332112123323342222321100000000001002002313200000000034021000000100010010100000100001001332000010001000000100000000000000000010000002023212331000000000110000000000001023241334341220000102134411100000002311230010002000100010123424344444444444444434444443344211000000000001000103100000000002323331110000010100000020001000000000420040013001202123434444443333333332233333334233333444544535666658 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCCSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MTPLCLNCSVLPGDLYPGGARNPMACNGSAARGHFDPEDLNLTDEALRLKYLGPQQTELFMPICATYLLIFVVGAVGNGLTCLVILRHKAMRTPTNYYLFSLAVSDLLVLLVGLPLELYEMWHNYPFLLGVGGCYFRTLLFEMVCLASVLNVTALSVERYVAVVHPLQARSMVTRAHVRRVLGAVWGLAMLCSLPNTSLHGIRQLHVPCRGPVPDSAVCMLVRPRALYNMVVQTTALLFFCLPMAIMSVLYLLIGLRLRRERLLLMQEAKGRGSAAARSRYTCRLQQHDRGRRQVTKMLFVLVVVFGICWAPFHADRVMWSVVSQWTDGLHLAFQHVHVISGIFFYLGSAANPVLYSLMSSRFRETFQEALCLGACCHRLRPRHSSHSLSRMTTGSTLCDVGSLGSWVHPLAGNDGPEAQQETDPS | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.28 | 0.25 | 0.70 | 2.65 | Download | -----------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM---TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--------GAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD--PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.25 | 0.23 | 0.73 | 3.88 | Download | -------------------------------------------------------KETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAED-WPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDYKGS----YLRICLLHPVQFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNIFEMLRIDEGGGSGGDGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPLLSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC----------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.29 | 0.25 | 0.70 | 3.23 | Download | -----------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--------GAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRL------------------LSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI--DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.25 | 0.20 | 0.73 | 1.57 | Download | -----------------------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK-GIET--------NPNNITCVLTKER--FGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETAAAAKGKNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC--N-QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR-------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.28 | 0.23 | 0.71 | 1.16 | Download | -------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRED------VDVIECSLQFPDDDDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEMIGRNTNGVIQTPNYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA----A----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.26 | 0.28 | 0.81 | 3.05 | Download | FRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--------GAVVCMLQFPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI--DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------------- | |||||||||||||||||||
| 7 | 4ib4 | 0.25 | 0.20 | 0.70 | 1.72 | Download | -------------------------------------------------------------WAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK-GIETN-P-------NNITCVLTKER--FGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNAKGTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC---NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCN---------------------------------------------------- | |||||||||||||||||||
| 8 | 4djhA | 0.27 | 0.23 | 0.71 | 4.50 | Download | -------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTT-MPFQSTVYLMNS-WPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR------EDVDVIECSLQFPSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEMLRIDEGLAKRVITTFRTGRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA--------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------------- | |||||||||||||||||||
| 9 | 4grvA | 0.30 | 0.23 | 0.69 | 2.81 | Download | -----------------------------------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR-KSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWHHPWAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNRS--ADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVIANKLTVMSGSV------------------------QALRHGVLVARAVVIAFVVCWLPYHVRRLMFCYISDEQWTLFDFYHYFYMLTNALAYASSAINPILYNLVSANFRQV------------------------------------------------------------ | |||||||||||||||||||
| 10 | 3zevA | 0.31 | 0.24 | 0.71 | 2.75 | Download | ----------------------------------------------PNSDLDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNIWVHHPWAFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNL--SGDGTHPGGLVCTPIVDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQP---------------------GRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDWTTFLFDFYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTLAC----------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
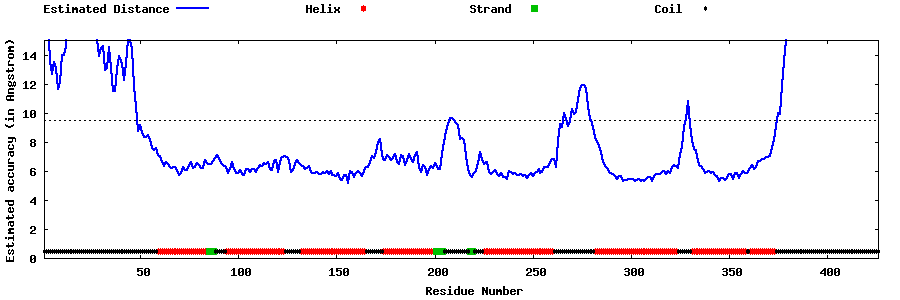
| Generated 3D models | Estimated local accuracy of models | ||
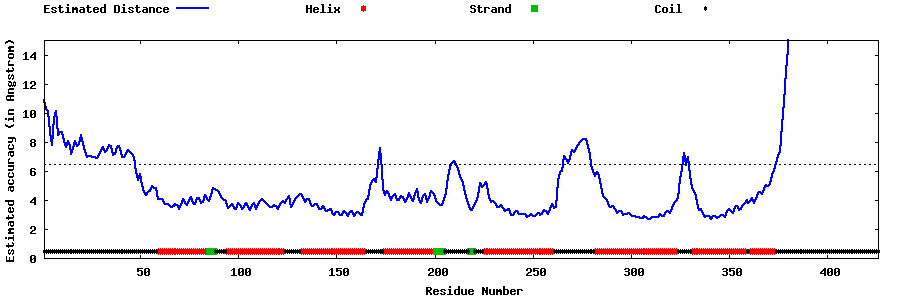 | |||
|
|
|||
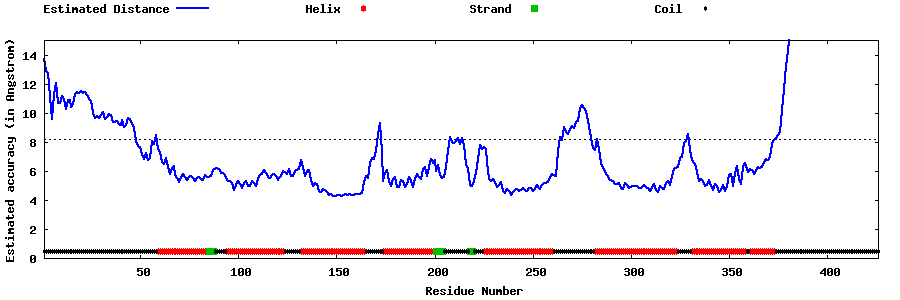 | |||
|
| |||
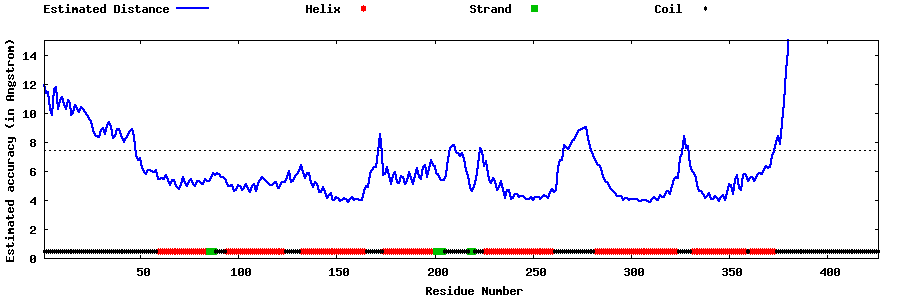 | |||
|
| |||
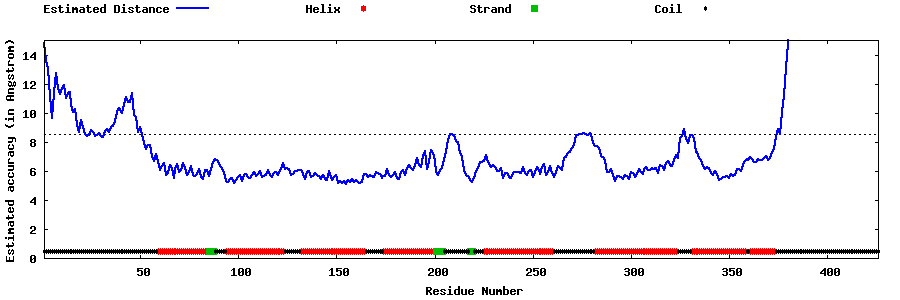 | |||
|
| |||
 | |||
|
| |||