

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MFLSSRMITSVSPSTSTNSSFLLTGFSGMEQQYPWLSIPFSSIYAMVLLGNCMVLHVIWTEPSLHQPMFYFLSMLALTDLCMGLSTVYTVLGILWGIIREISLDSCIAQSYFIHGLSFMESSVLLTMAFDRYIAICNPLRYSSILTNSRIIKIGLTIIGRSFFFITPPIICLKFFNYCHFHILSHSFCLHQDLLRLACSDIRFNSYYALMLVICILLLDAILILFSYILILKSVLAVASQEERHKLFQTCISHICAVLVFYIPIISLTMVHRFGKHLSPVAHVLIGNIYILFPPLMNPIIYSVKTQQIHTRMLRLFSLKRY | |
| CCCCHHHHHCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCC | |
| 977454440669997604001157998987789999999999999999999999999966887543289999999999999998559999999846998688888999999999999999999999981316751565446320888999999999999999999989999952899998947874321065688824783386899999999999998999999999999998078897788898551699999999999999999721457898993788999999862785536801116569999999999533669 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MFLSSRMITSVSPSTSTNSSFLLTGFSGMEQQYPWLSIPFSSIYAMVLLGNCMVLHVIWTEPSLHQPMFYFLSMLALTDLCMGLSTVYTVLGILWGIIREISLDSCIAQSYFIHGLSFMESSVLLTMAFDRYIAICNPLRYSSILTNSRIIKIGLTIIGRSFFFITPPIICLKFFNYCHFHILSHSFCLHQDLLRLACSDIRFNSYYALMLVICILLLDAILILFSYILILKSVLAVASQEERHKLFQTCISHICAVLVFYIPIISLTMVHRFGKHLSPVAHVLIGNIYILFPPLMNPIIYSVKTQQIHTRMLRLFSLKRY | |
| 732334303322334231220200000121320200011022022203321210000020141002000200010020020003002020000000213603140000001200310330110001002000000021010000003310010000002212011202101021031046200000001021001000230300111002003313332231032011100200020014601320110010000000003211321010010133101100001022100300330030000203300410030025468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCHHHHHCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCC MFLSSRMITSVSPSTSTNSSFLLTGFSGMEQQYPWLSIPFSSIYAMVLLGNCMVLHVIWTEPSLHQPMFYFLSMLALTDLCMGLSTVYTVLGILWGIIREISLDSCIAQSYFIHGLSFMESSVLLTMAFDRYIAICNPLRYSSILTNSRIIKIGLTIIGRSFFFITPPIICLKFFNYCHFHILSHSFCLHQDLLRLACSDIRFNSYYALMLVICILLLDAILILFSYILILKSVLAVASQEERHKLFQTCISHICAVLVFYIPIISLTMVHRFGKHLSPVAHVLIGNIYILFPPLMNPIIYSVKTQQIHTRMLRLFSLKRY | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.22 | 0.86 | 2.41 | Download | ----------------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.22 | 0.84 | 2.06 | Download | ------------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 3 | 4iaqA | 0.19 | 0.22 | 0.83 | 1.95 | Download | ---------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTG--RWTLVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLP-------PFFWRE-----------------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHL-----AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 4 | 4ib4 | 0.15 | 0.21 | 0.87 | 1.49 | Download | --------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGI--ET----NPNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAAISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR | |||||||||||||||||||
| 5 | 3uonA | 0.19 | 0.21 | 0.85 | 1.13 | Download | -----------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQF--IVGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKS----RREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---- | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.22 | 0.86 | 2.60 | Download | ------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.25 | 0.82 | 1.70 | Download | -------------------------------WKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIAAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH----LAIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 8 | 4buoA | 0.14 | 0.18 | 0.90 | 3.18 | Download | -----------------NSD---LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPIV------DTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRAVVIAFVVCWLPYHVRRLMFCYISDEQWTYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL----- | |||||||||||||||||||
| 9 | 5tgzA | 0.19 | 0.22 | 0.89 | 3.08 | Download | --------GGRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 10 | 4gpoA | 0.21 | 0.22 | 0.87 | 4.54 | Download | ----------------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
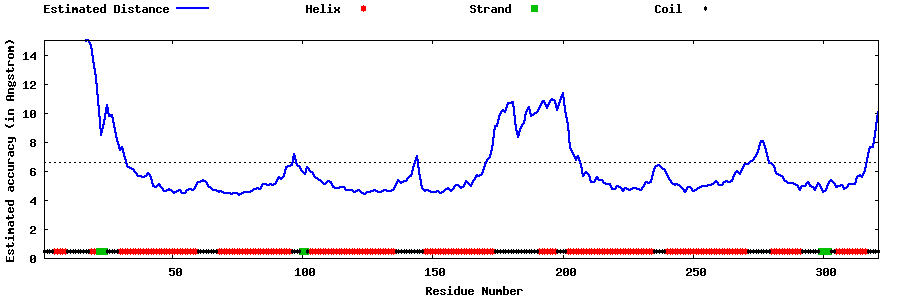
| Generated 3D models | Estimated local accuracy of models | ||
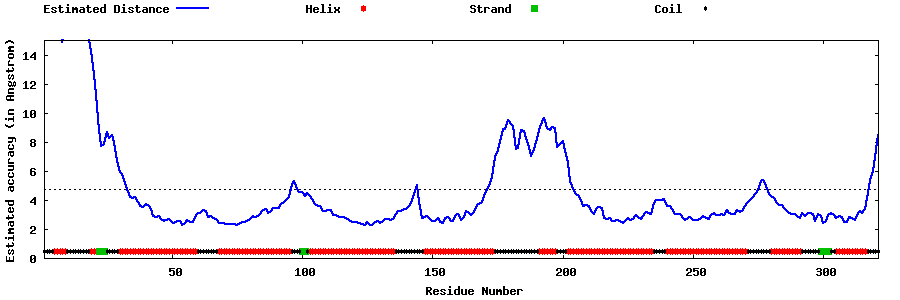 | |||
|
|
|||
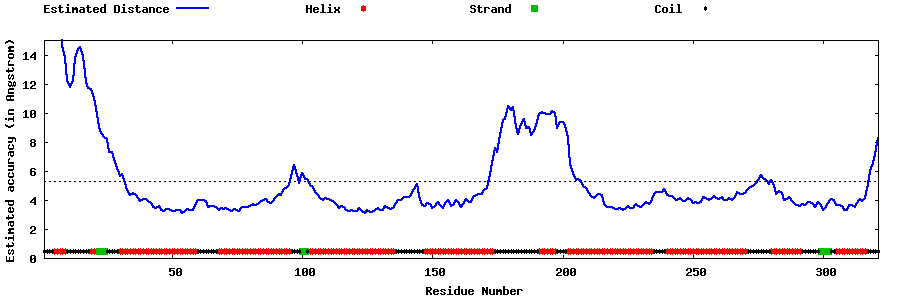 | |||
|
| |||
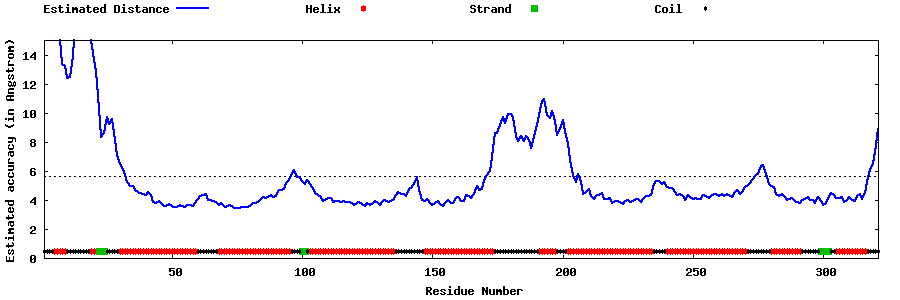 | |||
|
| |||
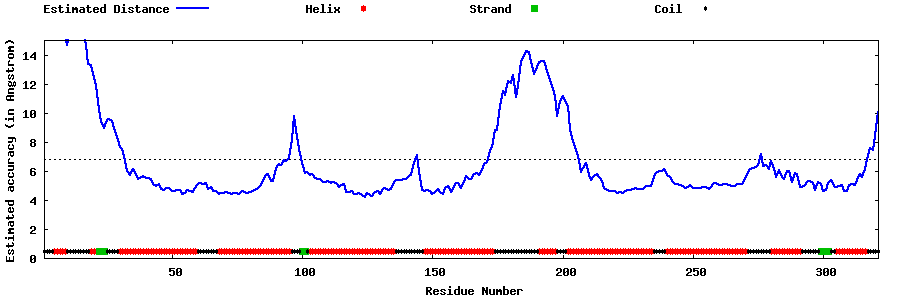 | |||
|
| |||
 | |||
|
| |||