

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MTNSSSTSTSSTTGGSLLLLCEEEESWAGRRIPVSLLYSGLAIGGTLANGMVIYLVSSFRKLQTTSNAFIVNGCAADLSVCALWMPQEAVLGLLPTGSAEPPADWDGAGGSYRLLRGGLLGLGLTVSLLSHCLVALNRYLLITRAPATYQALYQRRHTAGMLALSWALALGLVLLLPPWAPRPGAAPPRVHYPALLAAAALLAQTALLLHCYLGIVRRVRVSVKRVSVLNFHLLHQLPGCAAAAAAFPGAQHAPGPGGAAHPAQAQPLPPALHPRRAQRRLSGLSVLLLCCVFLLATQPLVWVSLASGFSLPVPWGVQAASWLLCCALSALNPLLYTWRNEEFRRSVRSVLPGVGDAAAAAVAATAVPAVSQAQLGTRAAGQHW | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSSCCHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHSSSHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCSSSCCCCCCCCCCCC | |
| 998887765778788756666766533789999999999999999998886067577668887487899999999999999999999999998435653346642206635998999999999999999999999872135876846545743288888867699999999999756225877888875316744220796996999999999999999999998776402200136653222212455444554456554332000146777778999998505520135689986999999999853589977999999999999997839999727999999999984615778998666788864334687876788899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MTNSSSTSTSSTTGGSLLLLCEEEESWAGRRIPVSLLYSGLAIGGTLANGMVIYLVSSFRKLQTTSNAFIVNGCAADLSVCALWMPQEAVLGLLPTGSAEPPADWDGAGGSYRLLRGGLLGLGLTVSLLSHCLVALNRYLLITRAPATYQALYQRRHTAGMLALSWALALGLVLLLPPWAPRPGAAPPRVHYPALLAAAALLAQTALLLHCYLGIVRRVRVSVKRVSVLNFHLLHQLPGCAAAAAAFPGAQHAPGPGGAAHPAQAQPLPPALHPRRAQRRLSGLSVLLLCCVFLLATQPLVWVSLASGFSLPVPWGVQAASWLLCCALSALNPLLYTWRNEEFRRSVRSVLPGVGDAAAAAVAATAVPAVSQAQLGTRAAGQHW | |
| 461433433333334434434344432201100001111310330331130000000103300100000010001000000000000000100133233223331110010000000000010000002000000100000002112414422132000000020233131311200100133232333100000002211331110002002200210131033134334423443443434444355444354454444444444344333233344121000000011102200311000000200102002000000001013000200000002045013001200112133464434343333234444344433644 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSSCCHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHSSSHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCSSSCCCCCCCCCCCC MTNSSSTSTSSTTGGSLLLLCEEEESWAGRRIPVSLLYSGLAIGGTLANGMVIYLVSSFRKLQTTSNAFIVNGCAADLSVCALWMPQEAVLGLLPTGSAEPPADWDGAGGSYRLLRGGLLGLGLTVSLLSHCLVALNRYLLITRAPATYQALYQRRHTAGMLALSWALALGLVLLLPPWAPRPGAAPPRVHYPALLAAAALLAQTALLLHCYLGIVRRVRVSVKRVSVLNFHLLHQLPGCAAAAAAFPGAQHAPGPGGAAHPAQAQPLPPALHPRRAQRRLSGLSVLLLCCVFLLATQPLVWVSLASGFSLPVPWGVQAASWLLCCALSALNPLLYTWRNEEFRRSVRSVLPGVGDAAAAAVAATAVPAVSQAQLGTRAAGQHW | |||||||||||||||||||||||||
| 1 | 4iaqA | 0.22 | 0.20 | 0.72 | 2.38 | Download | -------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVT---------GRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITD-AVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQAECVVNTHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRI---------------------------------------------IQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------------ | |||||||||||||||||||
| 2 | 4z34A | 0.17 | 0.20 | 0.88 | 4.18 | Download | -NEPQCFYNESIAFFYNRSGKHLATEWNTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRR----------LTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRM--QLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKAMKDNAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQDVLAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQIL--------------------------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.21 | 0.22 | 0.84 | 2.82 | Download | -------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWT---------LGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKR-TPKRAAVMIALVWVFSISISLPPFFWRQECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVRAAALDAQKATPDSPEMKDFRHKTTRNAYIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------------ | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.23 | 0.84 | 1.56 | Download | ------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEA-------MWP-LPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKK-PIQANQYNSRATAFIKITVVWLISIGIAIPVPIIETNPNTCLTFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLADNAAQVRAAADQKANEGKVKEAQQKTNAYIQKYLQTISNEQRAS-KVLGIVFFLFLLMWCPFFITNITLVLCDSTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---------------------------- | |||||||||||||||||||
| 5 | 3d4s | 0.17 | 0.22 | 0.82 | 1.18 | Download | -----------------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKM---------WTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITS-PFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDEGLRDTEGYYTIGILDKAIGRNTNGVITKEQQKSTGTWDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL------------------------------- | |||||||||||||||||||
| 6 | 4iaqA | 0.20 | 0.22 | 0.83 | 2.71 | Download | --------------------IYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWT---------LGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITD-AVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQASVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLEDNWETLNDNLKVINAAQVKDALTKMRAAAAAEQLKTTRNAYIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPI----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------------ | |||||||||||||||||||
| 7 | 4ib4 | 0.17 | 0.23 | 0.82 | 1.75 | Download | -------------------------------HWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMF--------EAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKP-IQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNPNTCVLDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVVKALTKMRAAADNEGKVKEAQAETTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNY----------------------------- | |||||||||||||||||||
| 8 | 4bvnA | 0.24 | 0.22 | 0.71 | 3.71 | Download | -------------------------LSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWL---------WGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITS-PFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRK-----------------------------------RKTS--------RVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRLVPKWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLLA-------------------------------- | |||||||||||||||||||
| 9 | 4iaqA | 0.22 | 0.20 | 0.72 | 2.52 | Download | -------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTG---------RWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITD-AVEYSAKRTPKRAAVMIALVWVFSISISLPPFFWRQAECVVNDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRI---------------------------------------------IQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMP----IHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------------ | |||||||||||||||||||
| 10 | 3zpqA | 0.25 | 0.23 | 0.71 | 2.37 | Download | ---------------------GAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGT---------WLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAIT-SPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQS-------------------------------------------------RVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRLLA-------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
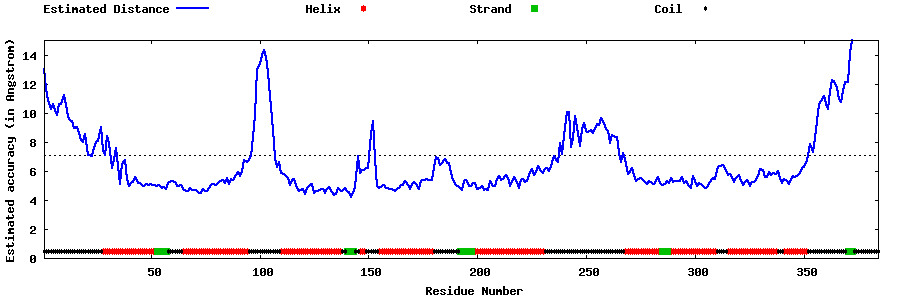
| Generated 3D models | Estimated local accuracy of models | ||
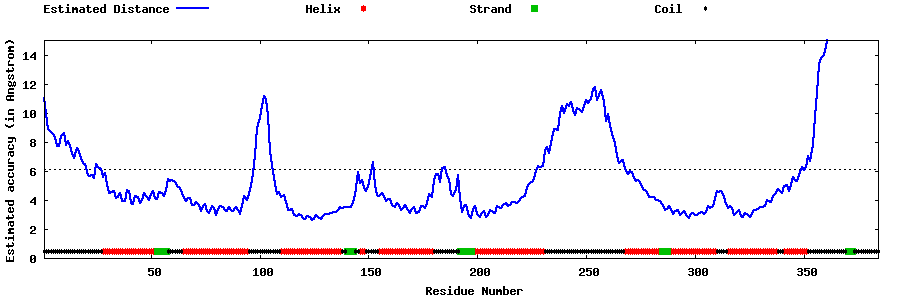 | |||
|
|
|||
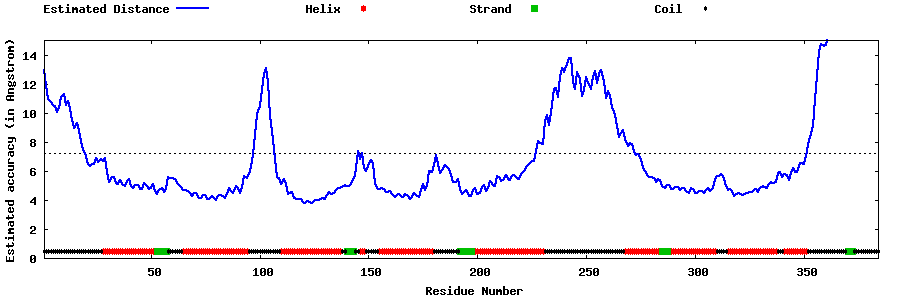 | |||
|
| |||
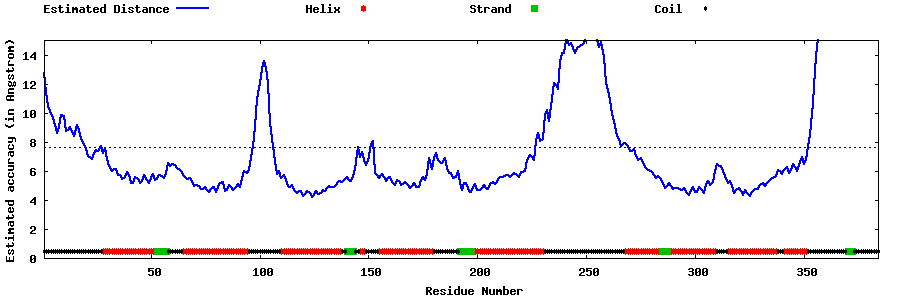 | |||
|
| |||
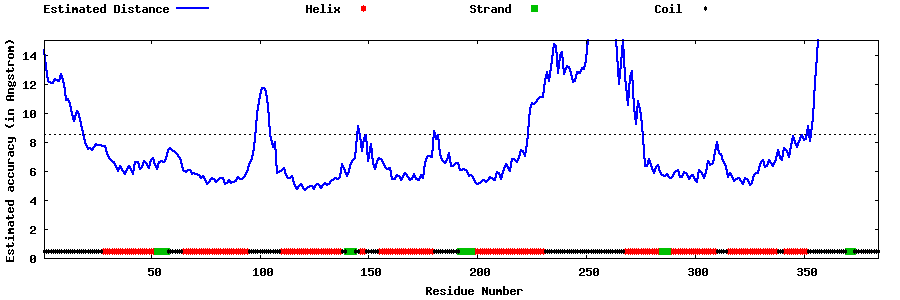 | |||
|
| |||
 | |||
|
| |||