

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MVFSAVLTAFHTGTSNTTFVVYENTYMNITLPPPFQHPDLSPLLRYSFETMAPTGLSSLTVNSTAVPTTPAAFKSLNLPLQITLSAIMIFILFVSFLGNLVVCLMVYQKAAMRSAINILLASLAFADMLLAVLNMPFALVTILTTRWIFGKFFCRVSAMFFWLFVIEGVAILLIISIDRFLIIVQRQDKLNPYRAKVLIAVSWATSFCVAFPLAVGNPDLQIPSRAPQCVFGYTTNPGYQAYVILISLISFFIPFLVILYSFMGILNTLRHNALRIHSYPEGICLSQASKLGLMSLQRPFQMSIDMGFKTRAFTTILILFAVFIVCWAPFTTYSLVATFSKHFYYQHNFFEISTWLLWLCYLKSALNPLIYYWRIKKFHDACLDMMPKSFKFLPQLPGHTKRRIRPSAVYVCGEHRTVV | |
| CSSSSSSCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 94788624766687776301113432445788888887745201367655788888777777888988888777778599999999999999999996898544466558988488999999999999999999999999999585748358887999999999999999999999872148753898787878888264999999999999999638400489987676204783003099999999999999999999999999999987742136554332222233465433330246688898999999999999999999999999999999826557870599999999999999998989999837999999999996054158888998766777898663048888679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MVFSAVLTAFHTGTSNTTFVVYENTYMNITLPPPFQHPDLSPLLRYSFETMAPTGLSSLTVNSTAVPTTPAAFKSLNLPLQITLSAIMIFILFVSFLGNLVVCLMVYQKAAMRSAINILLASLAFADMLLAVLNMPFALVTILTTRWIFGKFFCRVSAMFFWLFVIEGVAILLIISIDRFLIIVQRQDKLNPYRAKVLIAVSWATSFCVAFPLAVGNPDLQIPSRAPQCVFGYTTNPGYQAYVILISLISFFIPFLVILYSFMGILNTLRHNALRIHSYPEGICLSQASKLGLMSLQRPFQMSIDMGFKTRAFTTILILFAVFIVCWAPFTTYSLVATFSKHFYYQHNFFEISTWLLWLCYLKSALNPLIYYWRIKKFHDACLDMMPKSFKFLPQLPGHTKRRIRPSAVYVCGEHRTVV | |
| 41130322233333321011123333132213322334413321333233232332332322113242333323313000000000110200220331320000000002301000100020003001110000010000100273200130011000000000000000000000000000001134301230000000000110101000000002213344422201020123312200000000211320010002001100110142034134354434344444444444434443334133320000000000000000010000000000003302334001000000000002000300001001163004001400200142244144434333434333324443236 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CSSSSSSCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MVFSAVLTAFHTGTSNTTFVVYENTYMNITLPPPFQHPDLSPLLRYSFETMAPTGLSSLTVNSTAVPTTPAAFKSLNLPLQITLSAIMIFILFVSFLGNLVVCLMVYQKAAMRSAINILLASLAFADMLLAVLNMPFALVTILTTRWIFGKFFCRVSAMFFWLFVIEGVAILLIISIDRFLIIVQRQDKLNPYRAKVLIAVSWATSFCVAFPLAVGNPDLQIPSRAPQCVFGYTTNPGYQAYVILISLISFFIPFLVILYSFMGILNTLRHNALRIHSYPEGICLSQASKLGLMSLQRPFQMSIDMGFKTRAFTTILILFAVFIVCWAPFTTYSLVATFSKHFYYQHNFFEISTWLLWLCYLKSALNPLIYYWRIKKFHDACLDMMPKSFKFLPQLPGHTKRRIRPSAVYVCGEHRTVV | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.19 | 0.23 | 0.87 | 3.08 | Download | KATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLG--SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------------------------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------- | |||||||||||||||||||
| 2 | 4iaqA | 0.27 | 0.20 | 0.66 | 2.37 | Download | ---------------------------------------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEKRTPKRAAVMIALVWVFSISISLPPFFWRQASE-------CVVNTDH----ILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRII-------------------------QKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH---------LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------------------ | |||||||||||||||||||
| 3 | 5jqhA | 0.21 | 0.21 | 0.85 | 2.93 | Download | ILRNAKLKPVYDSLDAVRRAALINMVFQMGETGVAGFTNSLRMLQQKRWDEAAVNLAKSRWYNQTPNTGTWDAYAADEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCYETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQ-----------------------------LDKFALKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIR----KEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCLR------------------------------ | |||||||||||||||||||
| 4 | 5gliA | 0.18 | 0.23 | 0.77 | 4.12 | Download | -----------------------------------------------------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMDYKGSICLLHPVQKTAATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNIFEMLRIDEGGGSGGDEAEKRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNRLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC------------------------------- | |||||||||||||||||||
| 5 | 5unfA | 0.14 | 0.20 | 0.85 | 2.78 | Download | NAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKGPKKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFP----WQASYIVPLVWCMACLSSLPTFYFRDVRTIELGVNACIMAFPPEQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSY---------------------------GKNRITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMGVINAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR-------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.20 | 0.71 | 3.32 | Download | --------------------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------------------------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------- | |||||||||||||||||||
| 7 | 4n6hA | 0.21 | 0.23 | 0.87 | 3.17 | Download | KVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKLANEGKVKEAQAANAYIQKYLGSPGA-RSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------------------------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------------- | |||||||||||||||||||
| 8 | 3uon | 0.19 | 0.22 | 0.72 | 1.18 | Download | -----------------------------------------------------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGTVEDGECYIQFFS---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGHNAANTNGVITKTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIP----NTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------------------------- | |||||||||||||||||||
| 9 | 2ziy | 0.17 | 0.17 | 0.85 | 1.53 | Download | --------------------------------------------------DLRDNETWWYNPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAAMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNCSFDYISRDTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNA-----------KELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV---TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEIPGESSDAAPSADAAQMK | |||||||||||||||||||
| 10 | 4ib4 | 0.23 | 0.20 | 0.72 | 1.71 | Download | --------------------------------------------------------------------------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANSRATAFIKITVVWLISIGIAIPVIKGIETN---PNNITCVLTK---ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDRAADKEGQTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN-QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNY----------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
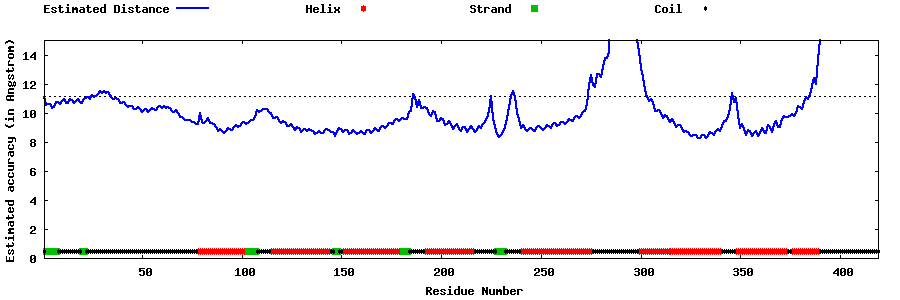
| Generated 3D models | Estimated local accuracy of models | ||
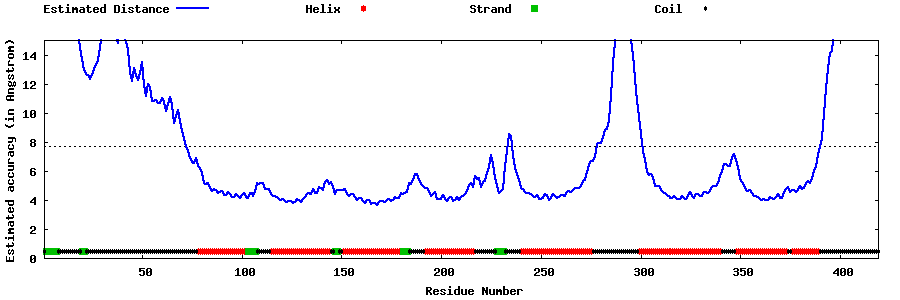 | |||
|
|
|||
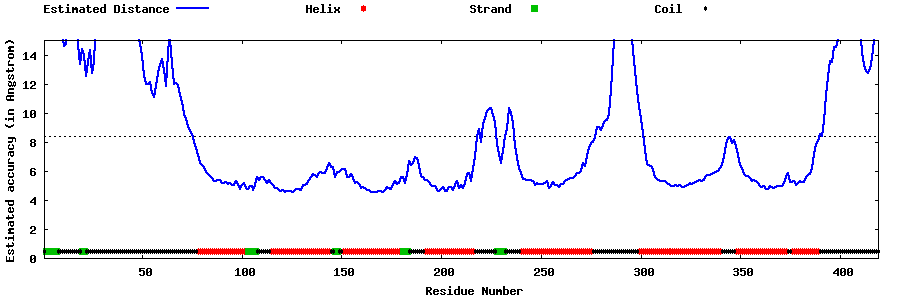 | |||
|
| |||
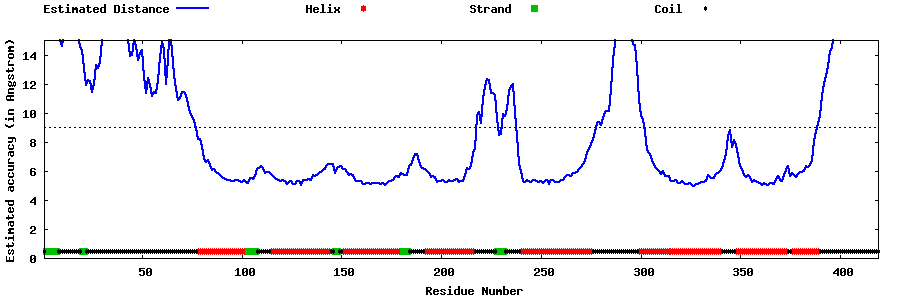 | |||
|
| |||
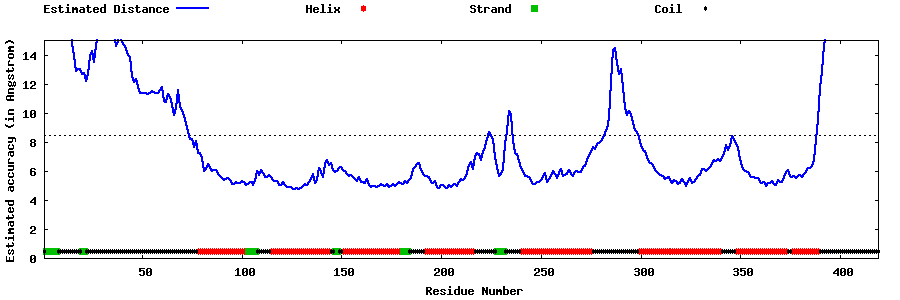 | |||
|
| |||
 | |||
|
| |||