

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MYNGSCCRIEGDTISQVMPPLLIVAFVLGALGNGVALCGFCFHMKTWKPSTVYLFNLAVADFLLMICLPFRTDYYLRRRHWAFGDIPCRVGLFTLAMNRAGSIVFLTVVAADRYFKVVHPHHAVNTISTRVAAGIVCTLWALVILGTVYLLLENHLCVQETAVSCESFIMESANGWHDIMFQLEFFMPLGIILFCSFKIVWSLRRRQQLARQARMKKATRFIMVVAIVFITCYLPSVSARLYFLWTVPSSACDPSVHGALHITLSFTYMNSMLDPLVYYFSSPSFPKFYNKLKICSLKPKQPGHSKTQRPEEMPISNLGRRSCISVANSFQSQSDGQWDPHIVEWH | |
| CCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHCCC | |
| 9899999876117898599999999999998899843400253788958899999999999999998899999995689998897569899999999999999999999997168874321011152556788888999999999999999800268559907997058526789999996001699999999999999999738888644320014889999999999998478999999999832876088999999999999999999998999993188888999999877546777777767788876777799888735778851478999995000369 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MYNGSCCRIEGDTISQVMPPLLIVAFVLGALGNGVALCGFCFHMKTWKPSTVYLFNLAVADFLLMICLPFRTDYYLRRRHWAFGDIPCRVGLFTLAMNRAGSIVFLTVVAADRYFKVVHPHHAVNTISTRVAAGIVCTLWALVILGTVYLLLENHLCVQETAVSCESFIMESANGWHDIMFQLEFFMPLGIILFCSFKIVWSLRRRQQLARQARMKKATRFIMVVAIVFITCYLPSVSARLYFLWTVPSSACDPSVHGALHITLSFTYMNSMLDPLVYYFSSPSFPKFYNKLKICSLKPKQPGHSKTQRPEEMPISNLGRRSCISVANSFQSQSDGQWDPHIVEWH | |
| 5615312424450111000220120023133232200100012334320000000000100000000010100000344421001001100001113111100100000021000000100202321333100000010100001001000001203467221000002254321021011021133112001200110011024254366544210000000000000000122101000010022340413300100111010000300011020100004501410240022224464444444544443444444442343354534444644433332618 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHCCC MYNGSCCRIEGDTISQVMPPLLIVAFVLGALGNGVALCGFCFHMKTWKPSTVYLFNLAVADFLLMICLPFRTDYYLRRRHWAFGDIPCRVGLFTLAMNRAGSIVFLTVVAADRYFKVVHPHHAVNTISTRVAAGIVCTLWALVILGTVYLLLENHLCVQETAVSCESFIMESANGWHDIMFQLEFFMPLGIILFCSFKIVWSLRRRQQLARQARMKKATRFIMVVAIVFITCYLPSVSARLYFLWTVPSSACDPSVHGALHITLSFTYMNSMLDPLVYYFSSPSFPKFYNKLKICSLKPKQPGHSKTQRPEEMPISNLGRRSCISVANSFQSQSDGQWDPHIVEWH | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.22 | 0.23 | 0.84 | 3.39 | Download | --------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAK-RTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEPPKVLCGVDHDKRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETR----STKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------------------------ | |||||||||||||||||||
| 2 | 4xnwA | 0.28 | 0.25 | 0.84 | 4.45 | Download | -SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVKNKTITCYDTTSDEYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.24 | 0.85 | 3.79 | Download | --SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 4 | 4djh | 0.25 | 0.24 | 0.80 | 1.54 | Download | ----------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYSFMICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-------AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.25 | 0.24 | 0.80 | 1.15 | Download | ----------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.23 | 0.23 | 0.84 | 3.41 | Download | ---------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAK-RTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYPKVLCGVDHDRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSA----RETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------------------------ | |||||||||||||||||||
| 7 | 4djh | 0.25 | 0.24 | 0.79 | 1.64 | Download | -----------PAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-------AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF---------------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.26 | 0.25 | 0.84 | 4.39 | Download | -------GSPGLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 9 | 5o9hA | 0.22 | 0.23 | 0.84 | 3.58 | Download | --------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFE-AKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEPPKVLCGVDHDKRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSA----RETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------------------------ | |||||||||||||||||||
| 10 | 4n6hA | 0.26 | 0.25 | 0.86 | 4.08 | Download | LGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKRRITRMVLVVVGAFVVCWAPIHIFVIVWTL-VDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
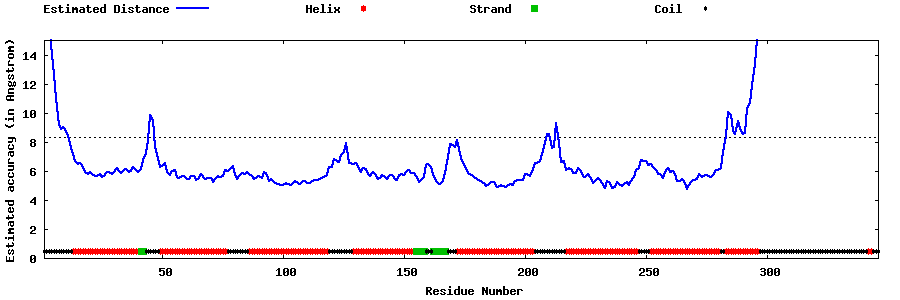
| Generated 3D models | Estimated local accuracy of models | ||
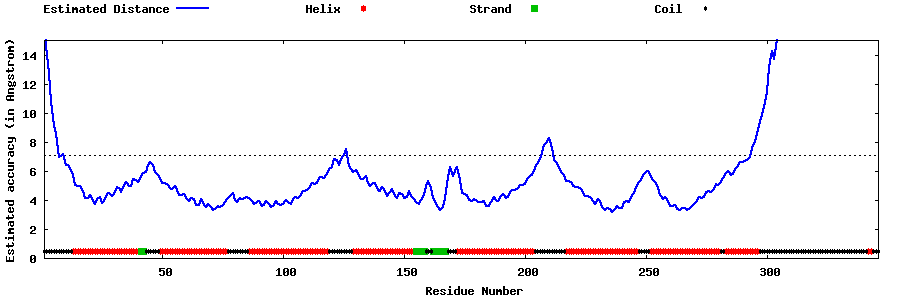 | |||
|
|
|||
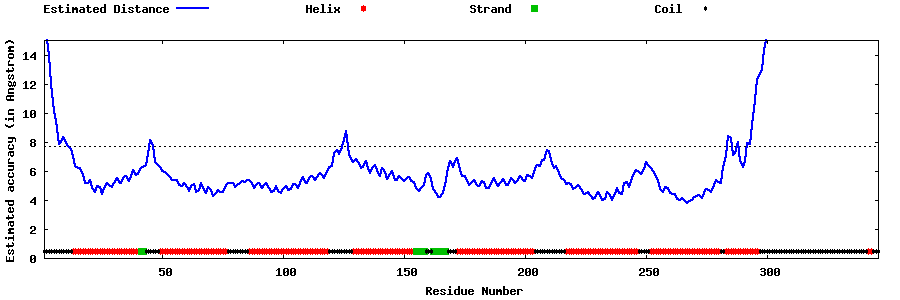 | |||
|
| |||
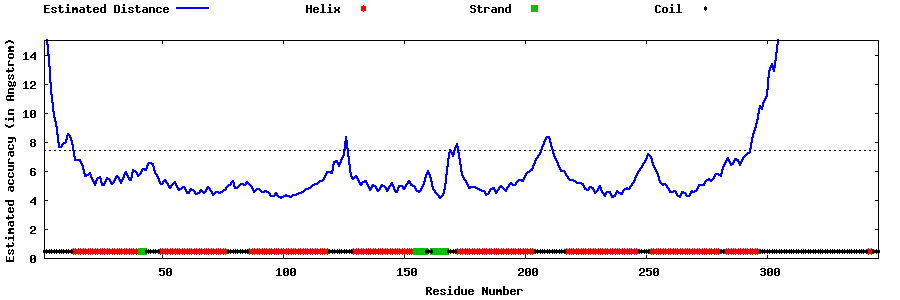 | |||
|
| |||
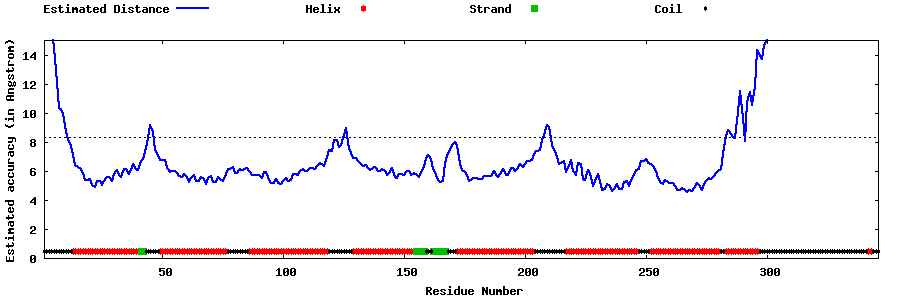 | |||
|
| |||
 | |||
|
| |||