

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MTAAIRRQRELSILPKVTLEAMNTTVMQGFNRSERCPRDTRIVQLVFPALYTVVFLTGILLNTLALWVFVHIPSSSTFIIYLKNTLVADLIMTLMLPFKILSDSHLAPWQLRAFVCRFSSVIFYETMYVGIVLLGLIAFDRFLKIIRPLRNIFLKKPVFAKTVSIFIWFFLFFISLPNTILSNKEATPSSVKKCASLKGPLGLKWHQMVNNICQFIFWTVFILMLVFYVVIAKKVYDSYRKSKSKDRKNNKKLEGKVFVVVAVFFVCFAPFHFARVPYTHSQTNNKTDCRLQNQLFIAKETTLFLAATNICMDPLIYIFLCKKFTEKLPCMQGRKTTASSQENHSSQTDNITLG | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCC | |
| 984324666678899998886767888899988778975127899899999999999999999988976516888785999999999999999970999999965899988965798999999999999999999999970575423032555767540100228999999999999999822266699937998258851178999999999888889999999999999999974166776543102246577777849999997149999999999998367898189999999999999999999999999999308899999999855667788878888888877897 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MTAAIRRQRELSILPKVTLEAMNTTVMQGFNRSERCPRDTRIVQLVFPALYTVVFLTGILLNTLALWVFVHIPSSSTFIIYLKNTLVADLIMTLMLPFKILSDSHLAPWQLRAFVCRFSSVIFYETMYVGIVLLGLIAFDRFLKIIRPLRNIFLKKPVFAKTVSIFIWFFLFFISLPNTILSNKEATPSSVKKCASLKGPLGLKWHQMVNNICQFIFWTVFILMLVFYVVIAKKVYDSYRKSKSKDRKNNKKLEGKVFVVVAVFFVCFAPFHFARVPYTHSQTNNKTDCRLQNQLFIAKETTLFLAATNICMDPLIYIFLCKKFTEKLPCMQGRKTTASSQENHSSQTDNITLG | |
| 644424454314323533343323133343433330434440110000230120023133232200100020353310000013002010000000101000003434320010011000011231111001000000310010001002023123220000000001100000010100022034475321000012366312011012211132133222200200000011013125545346444210000000000000110133101010010023033244041330021012101000030003100000000460141024004333444456444444664347 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCC MTAAIRRQRELSILPKVTLEAMNTTVMQGFNRSERCPRDTRIVQLVFPALYTVVFLTGILLNTLALWVFVHIPSSSTFIIYLKNTLVADLIMTLMLPFKILSDSHLAPWQLRAFVCRFSSVIFYETMYVGIVLLGLIAFDRFLKIIRPLRNIFLKKPVFAKTVSIFIWFFLFFISLPNTILSNKEATPSSVKKCASLKGPLGLKWHQMVNNICQFIFWTVFILMLVFYVVIAKKVYDSYRKSKSKDRKNNKKLEGKVFVVVAVFFVCFAPFHFARVPYTHSQTNNKTDCRLQNQLFIAKETTLFLAATNICMDPLIYIFLCKKFTEKLPCMQGRKTTASSQENHSSQTDNITLG | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.20 | 0.22 | 0.94 | 2.93 | Download | KLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-----DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.22 | 0.21 | 0.84 | 4.21 | Download | ------------------------------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYLRSYFYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYEEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLQTPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRL-------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.22 | 0.19 | 0.83 | 3.77 | Download | --------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLH-YTCSSHFPQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRM-----KEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------- | |||||||||||||||||||
| 4 | 4djh | 0.22 | 0.25 | 0.80 | 1.51 | Download | ---------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWDLFKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------------------- | |||||||||||||||||||
| 5 | 4xnv | 0.22 | 0.21 | 0.83 | 1.17 | Download | --------------------------------SFKCATKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYLSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKYTGVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR--------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.21 | 0.19 | 0.82 | 3.34 | Download | ----------------------------------QKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK-----EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------- | |||||||||||||||||||
| 7 | 4djh | 0.23 | 0.25 | 0.79 | 1.69 | Download | -----------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSWLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS----AA-------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF--------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.22 | 0.20 | 0.82 | 4.17 | Download | --------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEG-LHYTCSSHYSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK---------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.22 | 0.19 | 0.82 | 3.46 | Download | ----------------------------------PCQKINQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMK-----EEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.21 | 0.22 | 0.94 | 3.94 | Download | MRAAALDAQKATPLEDKSPDSRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD-----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
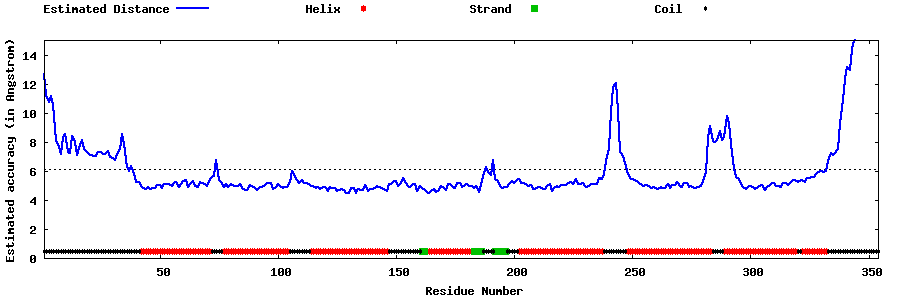
| Generated 3D models | Estimated local accuracy of models | ||
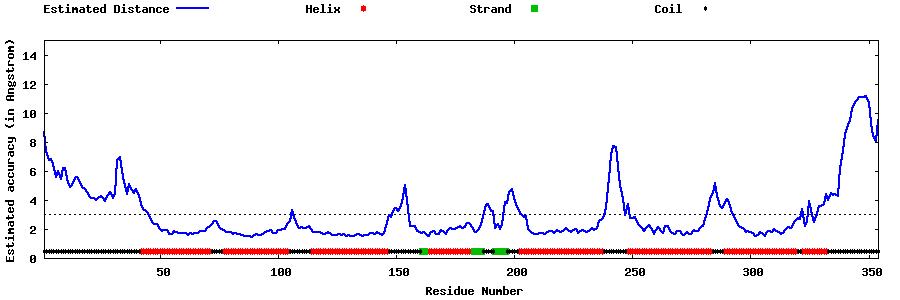 | |||
|
|
|||
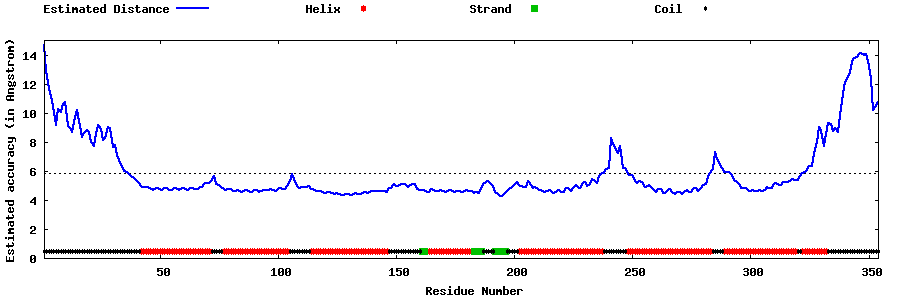 | |||
|
| |||
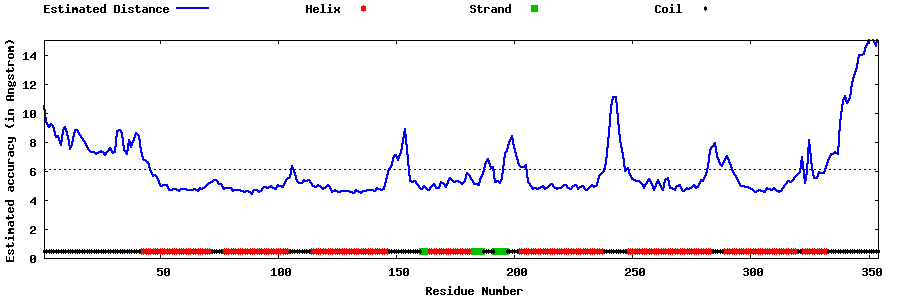 | |||
|
| |||
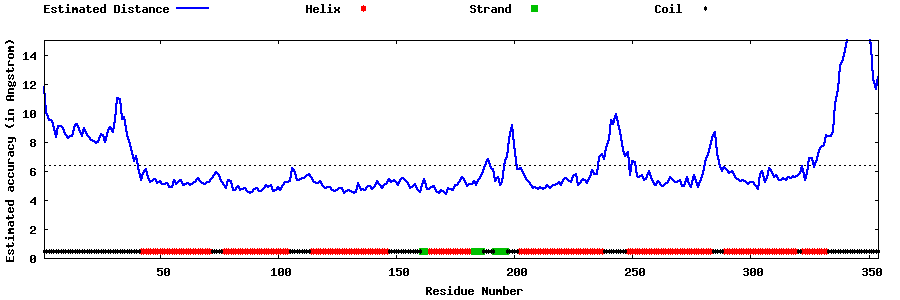 | |||
|
| |||
 | |||
|
| |||