

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MRMEDEDYNTSISYGDEYPDYLDSIVVLEDLSPLEARVTRIFLVVVYSIVCFLGILGNGLVIIIATFKMKKTVNMVWFLNLAVADFLFNVFLPIHITYAAMDYHWVFGTAMCKISNFLLIHNMFTSVFLLTIISSDRCISVLLPVWSQNHRSVRLAYMACMVIWVLAFFLSSPSLVFRDTANLHGKISCFNNFSLSTPGSSSWPTHSQMDPVGYSRHMVVTVTRFLCGFLVPVLIITACYLTIVCKLQRNRLAKTKKPFKIIVTIIITFFLCWCPYHTLNLLELHHTAMPGSVFSLGLPLATALAIANSCMNPILYVFMGQDFKKFKVALFSRLVNALSEDTGHSSYPSHRSFTKMSSMNERTSMNERETGML | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCSSSSSCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9887777777777787777666765566655533221899989999999999998778860123431677768999999999999999998299999997689886876689999999999999999999999999888865006402255515676778999999999982999987417449906882047866544224653210231257999999999999999999999999999999999535763466288999999999999979999999999963243158999999999999999985819999911686799999999988632565578888888877772538898887787555579 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MRMEDEDYNTSISYGDEYPDYLDSIVVLEDLSPLEARVTRIFLVVVYSIVCFLGILGNGLVIIIATFKMKKTVNMVWFLNLAVADFLFNVFLPIHITYAAMDYHWVFGTAMCKISNFLLIHNMFTSVFLLTIISSDRCISVLLPVWSQNHRSVRLAYMACMVIWVLAFFLSSPSLVFRDTANLHGKISCFNNFSLSTPGSSSWPTHSQMDPVGYSRHMVVTVTRFLCGFLVPVLIITACYLTIVCKLQRNRLAKTKKPFKIIVTIIITFFLCWCPYHTLNLLELHHTAMPGSVFSLGLPLATALAIANSCMNPILYVFMGQDFKKFKVALFSRLVNALSEDTGHSSYPSHRSFTKMSSMNERTSMNERETGML | |
| 6615344133333233323222333333443442323001000021012002302320110000020233110000000000100010000000100100353301001000120011133111100000000010000000000202322322101000000010000001000000203537422100020334344433243343244332202001111111101231122023310100010233444642000000000000010031211000001003314333101000000000012400310000000043014102300330240246444434444544334444444434444443247 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCSSSSSCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MRMEDEDYNTSISYGDEYPDYLDSIVVLEDLSPLEARVTRIFLVVVYSIVCFLGILGNGLVIIIATFKMKKTVNMVWFLNLAVADFLFNVFLPIHITYAAMDYHWVFGTAMCKISNFLLIHNMFTSVFLLTIISSDRCISVLLPVWSQNHRSVRLAYMACMVIWVLAFFLSSPSLVFRDTANLHGKISCFNNFSLSTPGSSSWPTHSQMDPVGYSRHMVVTVTRFLCGFLVPVLIITACYLTIVCKLQRNRLAKTKKPFKIIVTIIITFFLCWCPYHTLNLLELHHTAMPGSVFSLGLPLATALAIANSCMNPILYVFMGQDFKKFKVALFSRLVNALSEDTGHSSYPSHRSFTKMSSMNERTSMNERETGML | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.34 | 0.28 | 0.78 | 2.99 | Download | ---------------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEPPKVLCGVDHD-------------------KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.27 | 0.22 | 0.77 | 4.09 | Download | ----------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILIYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQ---------------YQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFF------------------------------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.31 | 0.27 | 0.78 | 3.93 | Download | ---------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKKTATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSP----------------SWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.30 | 0.27 | 0.74 | 1.53 | Download | -----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDD--------------YSW-WDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.30 | 0.27 | 0.74 | 1.16 | Download | -----------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDD---------------YSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA-----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.35 | 0.28 | 0.77 | 3.22 | Download | ----------------------------------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYPKVLCGVDHD-------------------KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.30 | 0.27 | 0.73 | 1.67 | Download | -------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIRYTKMKTATNIYIFNLALADALVTTTMPFQST-VYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDDY---S------------WWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF----------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.27 | 0.21 | 0.76 | 4.44 | Download | ----------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQ---------------YQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK--KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------------- | |||||||||||||||||||
| 9 | 5o9hA | 0.34 | 0.28 | 0.78 | 3.29 | Download | ---------------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEPPKVLCGVDHD-------------------KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.30 | 0.26 | 0.77 | 3.67 | Download | ------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHP----------------TWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
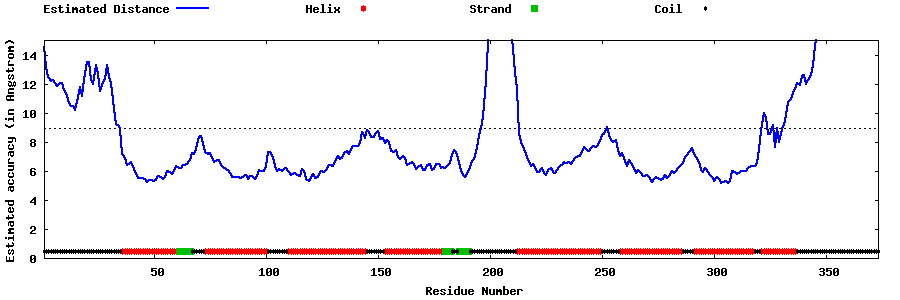
| Generated 3D models | Estimated local accuracy of models | ||
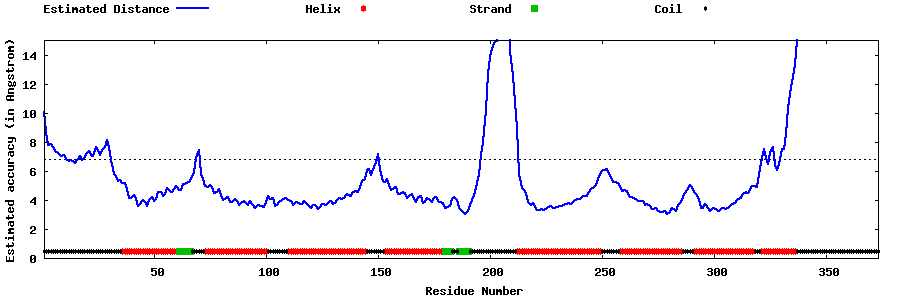 | |||
|
|
|||
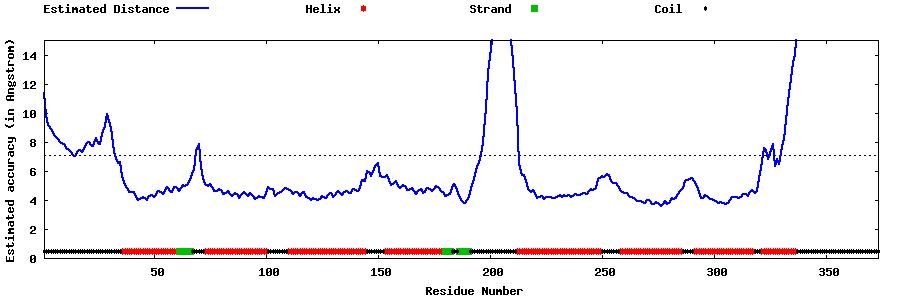 | |||
|
| |||
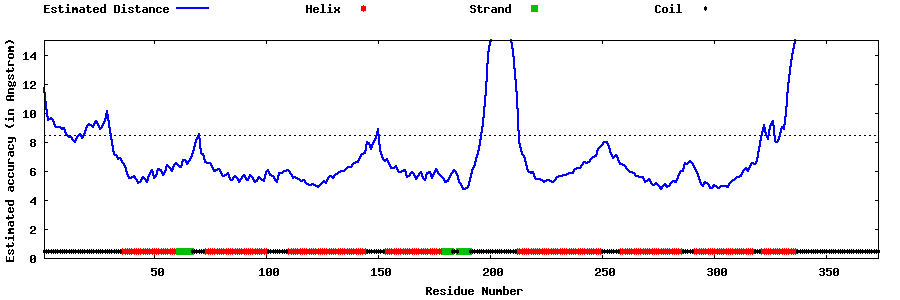 | |||
|
| |||
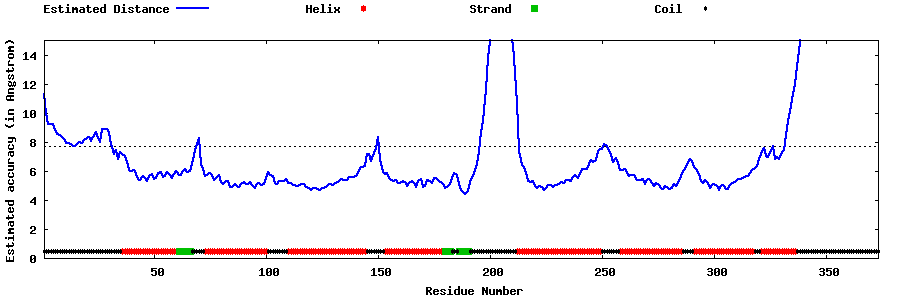 | |||
|
| |||
 | |||
|
| |||