

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MSVGAMKKGVGRAVGLGGGSGCQATEEDPLPNCGACAPGQGGRRWRLPQPAWVEGSSARLWEQATGTGWMDLEASLLPTGPNASNTSDGPDNLTSAGSPPRTGSISYINIIMPSVFGTICLLGIIGNSTVIFAVVKKSKLHWCNNVPDIFIINLSVVDLLFLLGMPFMIHQLMGNGVWHFGETMCTLITAMDANSQFTSTYILTAMAIDRYLATVHPISSTKFRKPSVATLVICLLWALSFISITPVWLYARLIPFPGGAVGCGIRLPNPDTDLYWFTLYQFFLAFALPFVVITAAYVRILQRMTSSVAPASQRSIRLRTKRVTRTAIAICLVFFVCWAPYYVLQLTQLSISRPTLTFVYLYNAAISLGYANSCLNPFVYIVLCETFRKRLVLSVKPAAQGQLRAVSNAQTADEERTESKGT | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC | |
| 97663012457788999886666776677888656799865656788985556775310015898887667310237789986657788777766667766442408999899999999999887799760212262568999989999999999999999998299999998389888887688899999999999999999999998588853206424475676888887999999999997999984668927992899624998278999999999999999999999999999999981768986411455541415224689999999997899999999993765248999999999999999997989999982898999999985254047766667788888778848999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MSVGAMKKGVGRAVGLGGGSGCQATEEDPLPNCGACAPGQGGRRWRLPQPAWVEGSSARLWEQATGTGWMDLEASLLPTGPNASNTSDGPDNLTSAGSPPRTGSISYINIIMPSVFGTICLLGIIGNSTVIFAVVKKSKLHWCNNVPDIFIINLSVVDLLFLLGMPFMIHQLMGNGVWHFGETMCTLITAMDANSQFTSTYILTAMAIDRYLATVHPISSTKFRKPSVATLVICLLWALSFISITPVWLYARLIPFPGGAVGCGIRLPNPDTDLYWFTLYQFFLAFALPFVVITAAYVRILQRMTSSVAPASQRSIRLRTKRVTRTAIAICLVFFVCWAPYYVLQLTQLSISRPTLTFVYLYNAAISLGYANSCLNPFVYIVLCETFRKRLVLSVKPAAQGQLRAVSNAQTADEERTESKGT | |
| 75233234012312035344414324556237434333364343342344332423433224423423213233323333332122133333233433144432230000000220120023023202200100000232444210000000000101100000000000000234420002001100100131001000000000000000000000202321221000000000100000001000000202537633110102023641210000011012012311200020000001101323444444444443200000000000001003110000000000223322000001000000002000000201000033004101400332154314634444444454454667 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC MSVGAMKKGVGRAVGLGGGSGCQATEEDPLPNCGACAPGQGGRRWRLPQPAWVEGSSARLWEQATGTGWMDLEASLLPTGPNASNTSDGPDNLTSAGSPPRTGSISYINIIMPSVFGTICLLGIIGNSTVIFAVVKKSKLHWCNNVPDIFIINLSVVDLLFLLGMPFMIHQLMGNGVWHFGETMCTLITAMDANSQFTSTYILTAMAIDRYLATVHPISSTKFRKPSVATLVICLLWALSFISITPVWLYARLIPFPGGAVGCGIRLPNPDTDLYWFTLYQFFLAFALPFVVITAAYVRILQRMTSSVAPASQRSIRLRTKRVTRTAIAICLVFFVCWAPYYVLQLTQLSISRPTLTFVYLYNAAISLGYANSCLNPFVYIVLCETFRKRLVLSVKPAAQGQLRAVSNAQTADEERTESKGT | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.25 | 0.29 | 0.93 | 3.19 | Download | VIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAE----QLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.30 | 0.29 | 0.70 | 3.61 | Download | -----------------------------------------------------------------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.23 | 0.29 | 0.93 | 3.86 | Download | --WETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 4 | 4n6hA | 0.29 | 0.24 | 0.71 | 3.81 | Download | ------------------------------------------------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 5 | 4n6hA | 0.24 | 0.29 | 0.93 | 3.46 | Download | VIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAE----QLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 6 | 4ib4 | 0.21 | 0.20 | 0.68 | 1.53 | Download | ------------------------------------------------------------------------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEK---KLQYATNYFLMSLAVADLLVGLFMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIET--NPNNITCVLTKER----FGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---------------------- | |||||||||||||||||||
| 7 | 4n6hA | 0.24 | 0.29 | 0.94 | 3.68 | Download | DNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKM---KTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG---------------------- | |||||||||||||||||||
| 8 | 4djh | 0.28 | 0.23 | 0.68 | 1.17 | Download | --------------------------------------------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMK---TATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREVDVIECSLQFPDDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.24 | 0.29 | 0.94 | 4.54 | Download | DNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK---TATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSWWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK----------------------PCG | |||||||||||||||||||
| 10 | 4djh | 0.28 | 0.23 | 0.67 | 1.73 | Download | -----------------------------------------------------------------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKT---ATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYSDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF-------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
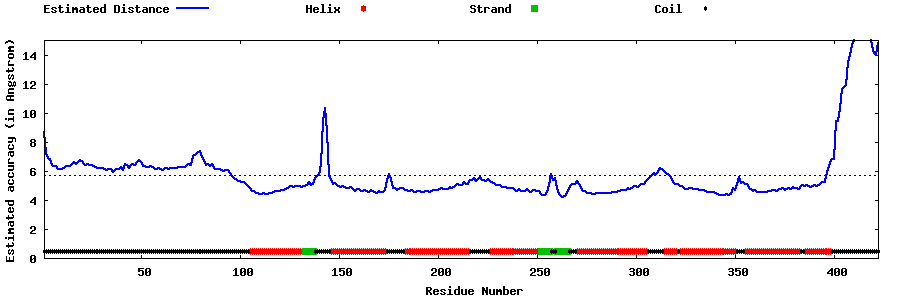
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||