

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MPSVSPAGPSAGAVPNATAVTTVRTNASGLEVPLFHLFARLDEELHGTFPGLWLALMAVHGAIFLAGLVLNGLALYVFCCRTRAKTPSVIYTINLVVTDLLVGLSLPTRFAVYYGARGCLRCAFPHVLGYFLNMHCSILFLTCICVDRYLAIVRPEGSRRCRQPACARAVCAFVWLAAGAVTLSVLGVTGSRPCCRVFALTVLEFLLPLLVISVFTGRIMCALSRPGLLHQGRQRRVRAMQLLLTVLIIFLVCFTPFHARQVAVALWPDMPHHTSLVVYHVAVTLSSLNSCMDPIVYCFVTSGFQATVRGLFGQHGEREPSSGDVVSMHRSSKGSGRHHILSAGPHALTQALANGPEA | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9999999876567888776777787788887765554445562002457427999999999999999988999678555067778688999999999999999979999999876565689999999999999999999999999998999982410011457267878889999999999865689828988617799999999999999999999999997367786533200035799999999999998289999999999971432589999999999999999989899898318999999999976315788777887765767888898761068997656555679999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MPSVSPAGPSAGAVPNATAVTTVRTNASGLEVPLFHLFARLDEELHGTFPGLWLALMAVHGAIFLAGLVLNGLALYVFCCRTRAKTPSVIYTINLVVTDLLVGLSLPTRFAVYYGARGCLRCAFPHVLGYFLNMHCSILFLTCICVDRYLAIVRPEGSRRCRQPACARAVCAFVWLAAGAVTLSVLGVTGSRPCCRVFALTVLEFLLPLLVISVFTGRIMCALSRPGLLHQGRQRRVRAMQLLLTVLIIFLVCFTPFHARQVAVALWPDMPHHTSLVVYHVAVTLSSLNSCMDPIVYCFVTSGFQATVRGLFGQHGEREPSSGDVVSMHRSSKGSGRHHILSAGPHALTQALANGPEA | |
| 7444336334343233313333334121323343242224134614442310100002301200230132122001000122343210000000001001000000001100010021121010011111210110000000000210000001002024223221000000001100000010100034321001100100101231120002000100010232434556453210000000000000000122100010010013123330010011001000130002100000000560141014003323435445454344443454444443343344434643663468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MPSVSPAGPSAGAVPNATAVTTVRTNASGLEVPLFHLFARLDEELHGTFPGLWLALMAVHGAIFLAGLVLNGLALYVFCCRTRAKTPSVIYTINLVVTDLLVGLSLPTRFAVYYGARGCLRCAFPHVLGYFLNMHCSILFLTCICVDRYLAIVRPEGSRRCRQPACARAVCAFVWLAAGAVTLSVLGVTGSRPCCRVFALTVLEFLLPLLVISVFTGRIMCALSRPGLLHQGRQRRVRAMQLLLTVLIIFLVCFTPFHARQVAVALWPDMPHHTSLVVYHVAVTLSSLNSCMDPIVYCFVTSGFQATVRGLFGQHGEREPSSGDVVSMHRSSKGSGRHHILSAGPHALTQALANGPEA | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.22 | 0.26 | 0.88 | 2.59 | Download | LVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMPFG-ELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVAVVCMLKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.23 | 0.21 | 0.75 | 3.61 | Download | ------------------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAWDFGNTMCQLLTGL-YFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTLHYTCSQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.26 | 0.24 | 0.77 | 3.37 | Download | ----------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMPFGE-LLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAGAVVCVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.20 | 0.20 | 0.75 | 1.55 | Download | ----------------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLVMPIALLTIMFEAMWVLC-PAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGPNNITCFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.30 | 0.25 | 0.73 | 1.16 | Download | ------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYSWPFGDVLCKIVLSI-DYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGDVIECSLQFCVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------------------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.26 | 0.24 | 0.77 | 3.04 | Download | ------------------------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLWPFGELLCKAVLSI-DYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.30 | 0.25 | 0.72 | 1.70 | Download | --------------------------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLWPFGDVLCKIVLSI-DYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTVIECSQFCVFIFAFVIPVLIIIVCYTLMILRLKSVRLLREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF--------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.27 | 0.24 | 0.77 | 4.28 | Download | ---------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLWPFGELLCKAVLSIDYY-NMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------------------------------- | |||||||||||||||||||
| 9 | 5o9hA | 0.24 | 0.21 | 0.75 | 3.18 | Download | ----------------------------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFE-AKRTINAIWFLNLAVADFLACLALPALFTSIV-PFGGAACSILP-SLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSAR-----ETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR----------------------------------- | |||||||||||||||||||
| 10 | 6b73A | 0.29 | 0.24 | 0.72 | 3.64 | Download | --------------------------------------------LGSISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLWPFGDVLCKIVLSIDY-YNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRSREKDR-NLRRITRLVLVVVAVFVVCWTPIHIFILVEALG-TSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKR---------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
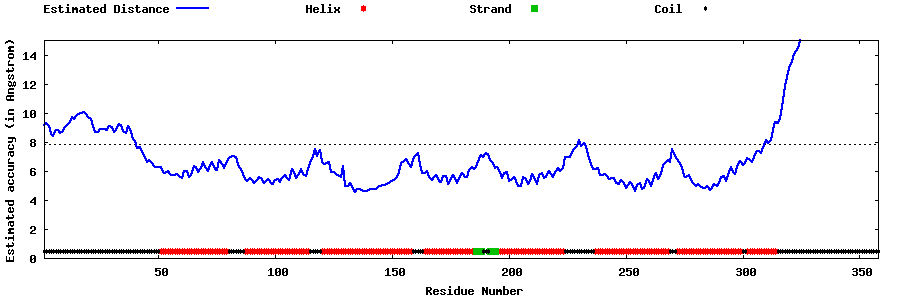
| Generated 3D models | Estimated local accuracy of models | ||
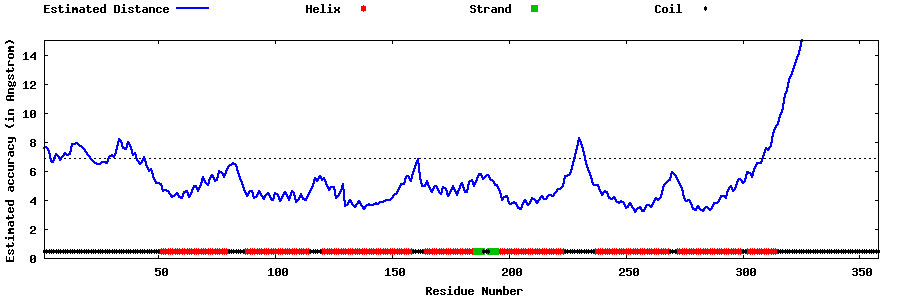 | |||
|
|
|||
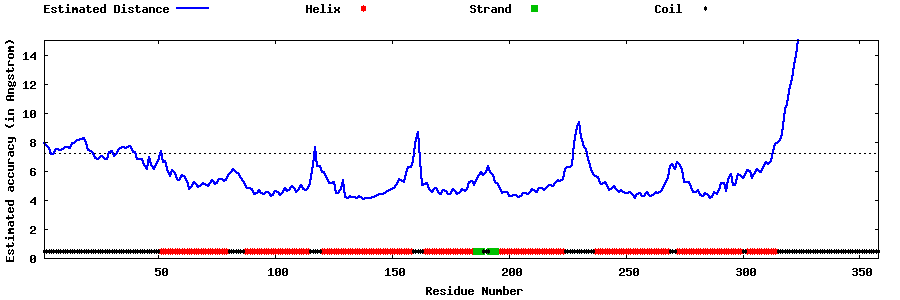 | |||
|
| |||
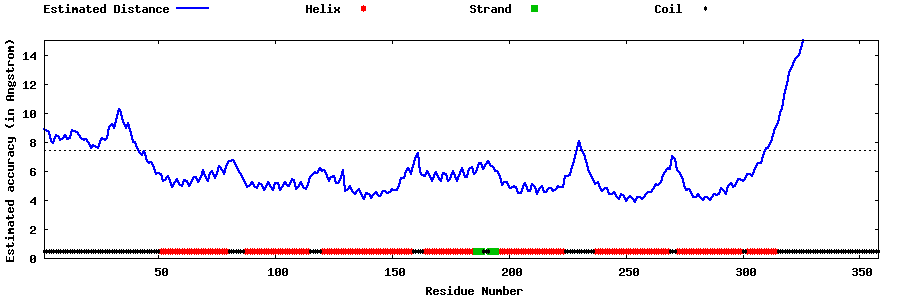 | |||
|
| |||
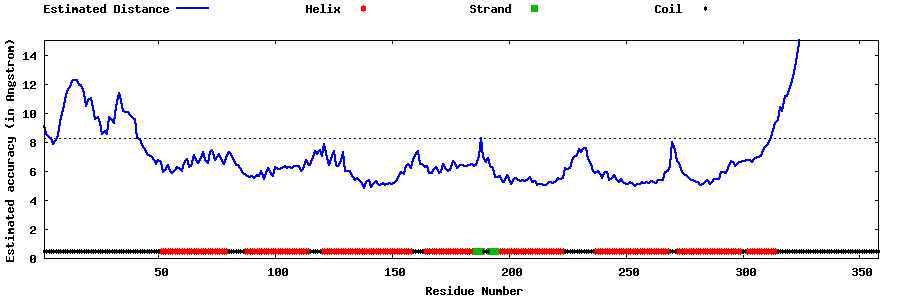 | |||
|
| |||
 | |||
|
| |||