

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDVTSQARGVGLEMYPGTAQPAAPNTTSPELNLSHPLLGTALANGTGELSEHQQYVIGLFLSCLYTIFLFPIGFVGNILILVVNISFREKMTIPDLYFINLAVADLILVADSLIEVFNLHERYYDIAVLCTFMSLFLQVNMYSSVFFLTWMSFDRYIALARAMRCSLFRTKHHARLSCGLIWMASVSATLVPFTAVHLQHTDEACFCFADVREVQWLEVTLGFIVPFAIIGLCYSLIVRVLVRAHRHRGLRPRRQKALRMILAVVLVFFVCWLPENVFISVHLLQRTQPGAAPCKQSFRHAHPLTGHIVNLAAFSNSCLNPLIYSFLGETFRDKLRLYIEQKTNLPALNRFCHAALKAVIPDSTEQSDVRFSSAV | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 987777778776518999999999888876566766677877888766554201447766799999999999997798742654307877878799999999999999999999999971898697888879999999999999999999999868999863521336451345211289999999999799883579846976899188888999999999999999999999999999705678654330244077763435489898773999999999999734677736899999999999999999999999999999974899999999997301589976766767666778999878777747879 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MDVTSQARGVGLEMYPGTAQPAAPNTTSPELNLSHPLLGTALANGTGELSEHQQYVIGLFLSCLYTIFLFPIGFVGNILILVVNISFREKMTIPDLYFINLAVADLILVADSLIEVFNLHERYYDIAVLCTFMSLFLQVNMYSSVFFLTWMSFDRYIALARAMRCSLFRTKHHARLSCGLIWMASVSATLVPFTAVHLQHTDEACFCFADVREVQWLEVTLGFIVPFAIIGLCYSLIVRVLVRAHRHRGLRPRRQKALRMILAVVLVFFVCWLPENVFISVHLLQRTQPGAAPCKQSFRHAHPLTGHIVNLAAFSNSCLNPLIYSFLGETFRDKLRLYIEQKTNLPALNRFCHAALKAVIPDSTEQSDVRFSSAV | |
| 763434444241423233243233433223211223234332324334246334200000002210110021023202100100011333210000000000100110000000000000310100110121010013211110000000023133000000020232132210100000011000000100000020353741010103142220010010122112102300000001013134455445321000000000000001002300000001002303234232333132001001000100012100310000000043014101300242243433433344344444453334344544437 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDVTSQARGVGLEMYPGTAQPAAPNTTSPELNLSHPLLGTALANGTGELSEHQQYVIGLFLSCLYTIFLFPIGFVGNILILVVNISFREKMTIPDLYFINLAVADLILVADSLIEVFNLHERYYDIAVLCTFMSLFLQVNMYSSVFFLTWMSFDRYIALARAMRCSLFRTKHHARLSCGLIWMASVSATLVPFTAVHLQHTDEACFCFADVREVQWLEVTLGFIVPFAIIGLCYSLIVRVLVRAHRHRGLRPRRQKALRMILAVVLVFFVCWLPENVFISVHLLQRTQPGAAPCKQSFRHAHPLTGHIVNLAAFSNSCLNPLIYSFLGETFRDKLRLYIEQKTNLPALNRFCHAALKAVIPDSTEQSDVRFSSAV | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.23 | 0.21 | 0.77 | 2.97 | Download | -----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEK---KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN---CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.23 | 0.21 | 0.77 | 4.09 | Download | -----------------------------------------------CQKINVKQIAARLLPPLYSL-VFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN---CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.24 | 0.24 | 0.77 | 3.54 | Download | ----------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMEWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD--------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.20 | 0.75 | 1.52 | Download | -----------------------------------------------EEQGNKLH-----WAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALTIEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIGIET--NPNNITCVLTKERFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN---------QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.23 | 0.20 | 0.75 | 1.16 | Download | ----------------------------------------------------VKQIGAQLLPPLYS-LVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAANEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE-DSVYVCGPYFPRHTIMRNILGLVLPLLIMVICYSGISRASKSRINIYPPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLS-NCE--STSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF------------------------------------ | |||||||||||||||||||
| 6 | 4mbsA | 0.23 | 0.21 | 0.76 | 3.14 | Download | -------------------------------------------------QKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE---KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN---NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.22 | 0.22 | 0.71 | 1.68 | Download | ------------------------------------------------------------FIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFMNLYTLTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVEDGECYIQFFNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP----C--------IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.23 | 0.21 | 0.75 | 4.28 | Download | ----------------------------------------------PCQKINVKQIAARLLPPLY-SLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFQTLKIVILGLVLPLLVMVICYSGILKTLLREKKRHRD-------VRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN---CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.23 | 0.21 | 0.77 | 3.29 | Download | -----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE---KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNN---CSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.24 | 0.22 | 0.75 | 3.62 | Download | ------------------------------------------GSHSLPQTGSPS-MVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET---------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
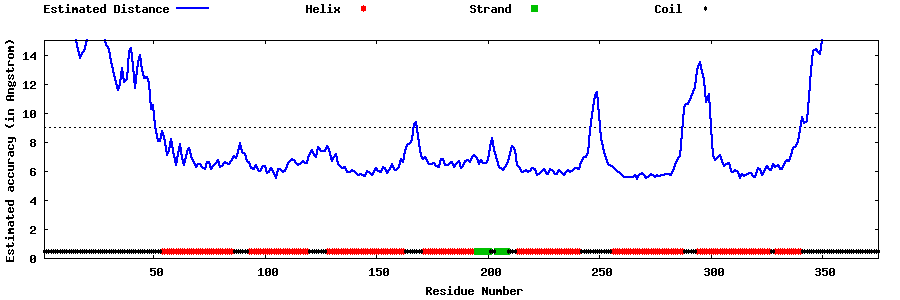
| Generated 3D models | Estimated local accuracy of models | ||
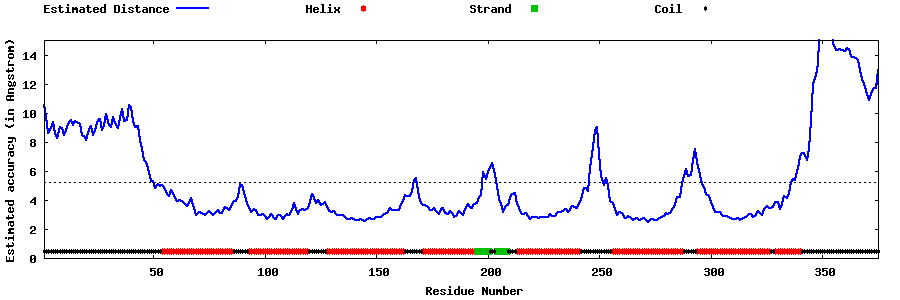 | |||
|
|
|||
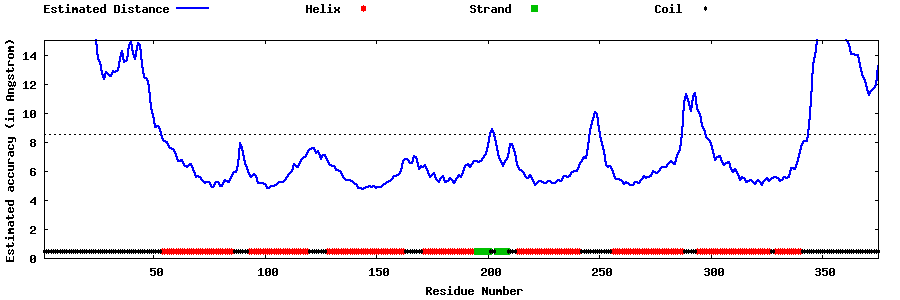 | |||
|
| |||
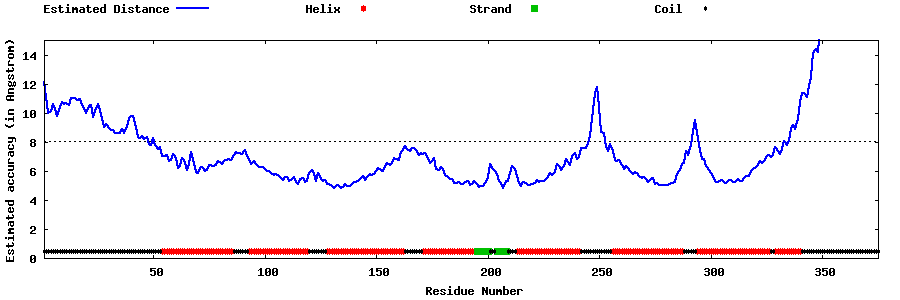 | |||
|
| |||
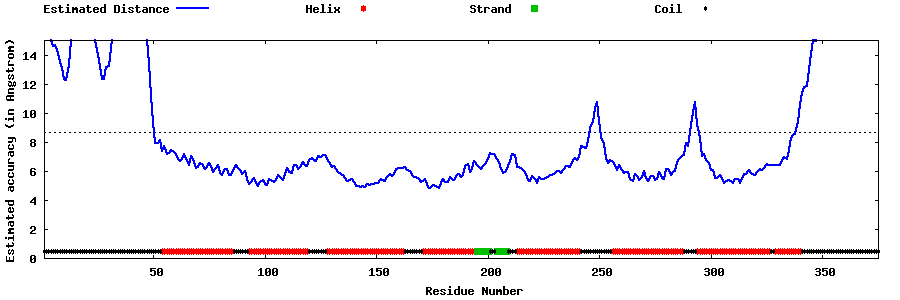 | |||
|
| |||
 | |||
|
| |||