

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MEVSLNHPASNTTSTKNNNSAFFYFESCQPPSPALLLLCIAYTVVLIVGLFGNLSLIIIIFKKQRKAQNFTSILIANLSLSDTLVCVMCIHFTIIYTLMDHWIFGDTMCRLTSYVQSVSISVSIFSLVFTAVERYQLIVNPRGWKPSVTHAYWGITLIWLFSLLLSIPFFLSYHLTDEPFRNLSLPTDLYTHQVACVENWPSKKDRLLFTTSLFLLQYFVPLGFILICYLKIVICLRRRNAKVDKKKENEGRLNENKRINTMLISIVVTFGACWLPRISSMSSLTGIMRC | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHCCHHHSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCCCCCCSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCC | |
| 99756799888778878888765554556762999999999998788850167870887777588878599999999999999999994699999998276036436898799999999999999999999982503421687633264688715599999999999999887799604777544445578873560469977899999999999999999999999999999998053666662166999999875899999999999999999999999999656669 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MEVSLNHPASNTTSTKNNNSAFFYFESCQPPSPALLLLCIAYTVVLIVGLFGNLSLIIIIFKKQRKAQNFTSILIANLSLSDTLVCVMCIHFTIIYTLMDHWIFGDTMCRLTSYVQSVSISVSIFSLVFTAVERYQLIVNPRGWKPSVTHAYWGITLIWLFSLLLSIPFFLSYHLTDEPFRNLSLPTDLYTHQVACVENWPSKKDRLLFTTSLFLLQYFVPLGFILICYLKIVICLRRRNAKVDKKKENEGRLNENKRINTMLISIVVTFGACWLPRISSMSSLTGIMRC | |
| 55432333342323333443323324424233001000021113002202321320010000123402100010020003002110200010101110253011130022001101110000002000000100020001002433333200000000111010001010001203434345342333334320101030127403200000103321331010002000100110233434354454344234432100000000000100033313011001132357 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHCCHHHSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCCCCCCSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCC MEVSLNHPASNTTSTKNNNSAFFYFESCQPPSPALLLLCIAYTVVLIVGLFGNLSLIIIIFKKQRKAQNFTSILIANLSLSDTLVCVMCIHFTIIYTLMDHWIFGDTMCRLTSYVQSVSISVSIFSLVFTAVERYQLIVNPRGWKPSVTHAYWGITLIWLFSLLLSIPFFLSYHLTDEPFRNLSLPTDLYTHQVACVENWPSKKDRLLFTTSLFLLQYFVPLGFILICYLKIVICLRRRNAKVDKKKENEGRLNENKRINTMLISIVVTFGACWLPRISSMSSLTGIMRC | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.22 | 0.26 | 0.86 | 2.33 | Download | ----------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY-TKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG------------AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDID | |||||||||||||||||||
| 2 | 1gzmA | 0.20 | 0.22 | 0.94 | 3.61 | Download | MNGTEGPPFSNKTGVVRSPFEAPQ--YYLAEPWQFSMLAAYMFLLIMLGFPINF-LTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEG-----------MQCSCGIDYYTETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESA---TTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGS | |||||||||||||||||||
| 3 | 4n6hA | 0.23 | 0.26 | 0.86 | 3.61 | Download | ----------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY-TKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDID | |||||||||||||||||||
| 4 | 4djh | 0.23 | 0.28 | 0.86 | 1.51 | Download | ------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMF-VIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE----------DVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIPAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA | |||||||||||||||||||
| 5 | 4djh | 0.23 | 0.28 | 0.86 | 1.13 | Download | ------------------------------SPAIPVIITAVYSVVFVVGLVGNS-LVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRED----------VDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNTPNRREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.26 | 0.86 | 2.81 | Download | ------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLV-MFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSG----SKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDID | |||||||||||||||||||
| 7 | 3uon | 0.18 | 0.24 | 0.67 | 1.68 | Download | ----------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIK-VNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRT---------VEDGECYIQFFSN---AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINI------------------------------------------------ | |||||||||||||||||||
| 8 | 1u19A | 0.21 | 0.22 | 0.95 | 3.54 | Download | MNGNFYVPFSNKTGV--VRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPIN-FLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYTPH----EETN----NESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQESA---TTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGS | |||||||||||||||||||
| 9 | 4n6hA | 0.22 | 0.26 | 0.86 | 2.60 | Download | ----------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVL-VMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD------------GAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDID | |||||||||||||||||||
| 10 | 5c1mA | 0.22 | 0.24 | 0.87 | 3.44 | Download | -------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFL-VMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQ------------GSIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS----KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIP | |||||||||||||||||||
| ||||||||||||||||||||||||||
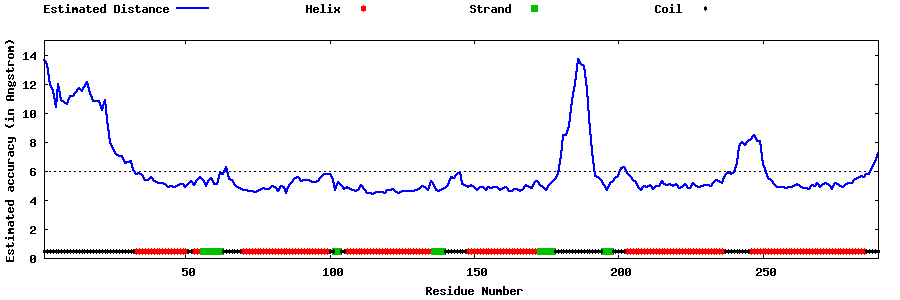
| Generated 3D models | Estimated local accuracy of models | ||
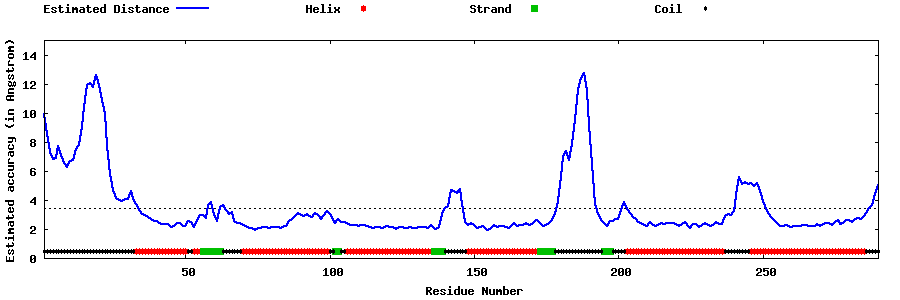 | |||
|
|
|||
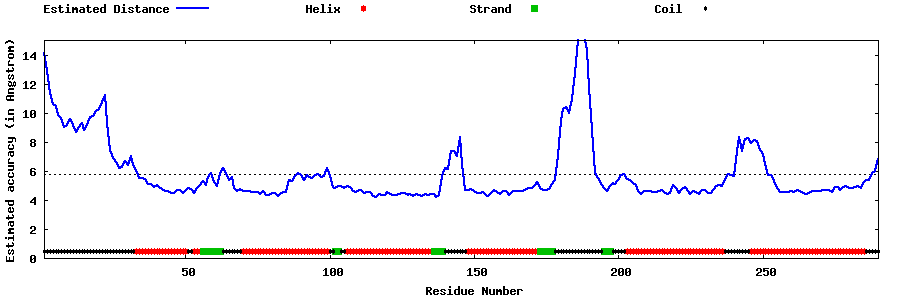 | |||
|
| |||
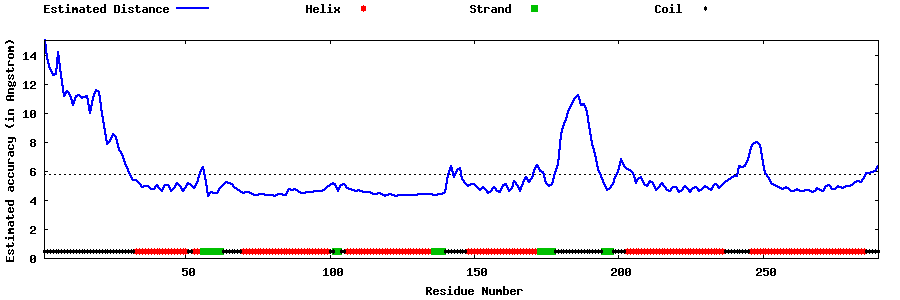 | |||
|
| |||
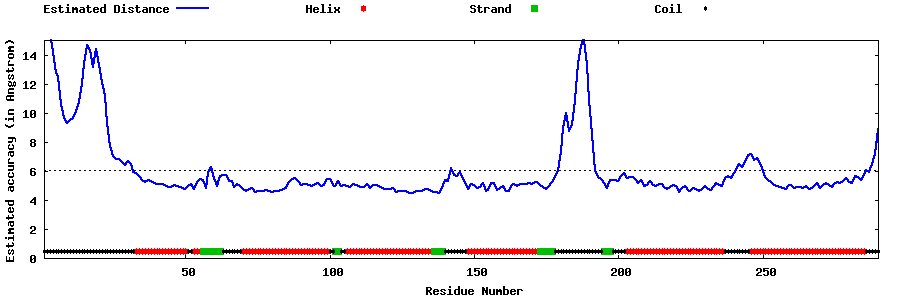 | |||
|
| |||
 | |||
|
| |||