

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MMPFCHNIINISCVKNNWSNDVRASLYSLMVLIILTTLVGNLIVIVSISHFKQLHTPTNWLIHSMATVDFLLGCLVMPYSMVRSAEHCWYFGEVFCKIHTSTDIMLSSASIFHLSFISIDRYYAVCDPLRYKAKMNILVICVMIFISWSVPAVFAFGMIFLELNFKGAEEIYYKHVHCRGGCSVFFSKISGVLTFMTSFYIPGSIMLCVYYRIYLIAKEQARLISDANQKLQIGLEMKNGISQSKERKAVKTLGIVMGVFLICWCPFFICTVMDPFLHYIIPPTLNDVLIWFGYLNSTFNPMVYAFFYPWFRKALKMMLFGKIFQKDSSRCKLFLELSS | |
| CCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHSSHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCC | |
| 988886877888899998878999999999999999999799853324545888854799999999999999999999999999818502764899999999999999999999999998788513414484320499999999999999999999999962644677666765566789884102109999999999999999999999999999987652111123433210002346787642333211238999999998899999999986388672899999999999998387999995789999999999486668999876344334688 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MMPFCHNIINISCVKNNWSNDVRASLYSLMVLIILTTLVGNLIVIVSISHFKQLHTPTNWLIHSMATVDFLLGCLVMPYSMVRSAEHCWYFGEVFCKIHTSTDIMLSSASIFHLSFISIDRYYAVCDPLRYKAKMNILVICVMIFISWSVPAVFAFGMIFLELNFKGAEEIYYKHVHCRGGCSVFFSKISGVLTFMTSFYIPGSIMLCVYYRIYLIAKEQARLISDANQKLQIGLEMKNGISQSKERKAVKTLGIVMGVFLICWCPFFICTVMDPFLHYIIPPTLNDVLIWFGYLNSTFNPMVYAFFYPWFRKALKMMLFGKIFQKDSSRCKLFLELSS | |
| 714011622232134662330110000011121023033312000000011330100000000000200110000002210010024230113101300000001000000300000000000000200304331223000000000121122202100001032443443234333224201010233002200320132002000200210010013203324425433334323332223333331032000000001000200000000200052101300000000201320120000000104301300120000200344133130145468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCHHHHHHHHHHHHHHHSSHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCC MMPFCHNIINISCVKNNWSNDVRASLYSLMVLIILTTLVGNLIVIVSISHFKQLHTPTNWLIHSMATVDFLLGCLVMPYSMVRSAEHCWYFGEVFCKIHTSTDIMLSSASIFHLSFISIDRYYAVCDPLRYKAKMNILVICVMIFISWSVPAVFAFGMIFLELNFKGAEEIYYKHVHCRGGCSVFFSKISGVLTFMTSFYIPGSIMLCVYYRIYLIAKEQARLISDANQKLQIGLEMKNGISQSKERKAVKTLGIVMGVFLICWCPFFICTVMDPFLHYIIPPTLNDVLIWFGYLNSTFNPMVYAFFYPWFRKALKMMLFGKIFQKDSSRCKLFLELSS | |||||||||||||||||||||||||
| 1 | 3sn6R | 0.34 | 0.29 | 0.84 | 2.87 | Download | -------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCY----AEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC------------------- | |||||||||||||||||||
| 2 | 4amjA | 0.35 | 0.31 | 0.86 | 4.67 | Download | ------------------SQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQ-ALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRKIDRASKRKT------SRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLL-------------------- | |||||||||||||||||||
| 3 | 3sn6R | 0.34 | 0.29 | 0.84 | 3.57 | Download | -------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCYAE----ETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEG------------RCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC------------------- | |||||||||||||||||||
| 4 | 3d4s | 0.32 | 0.30 | 0.87 | 1.56 | Download | ---------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQ----EAINCYTCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMARNTNGVIGTWDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL------------------ | |||||||||||||||||||
| 5 | 3d4s | 0.32 | 0.30 | 0.88 | 1.19 | Download | ---------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQEAINCY-AEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEKAIGRNTNGGTWDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL------------------ | |||||||||||||||||||
| 6 | 3sn6R | 0.34 | 0.29 | 0.84 | 3.12 | Download | -------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCYAE----ETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC------------------- | |||||||||||||||||||
| 7 | 3uon | 0.26 | 0.28 | 0.87 | 1.73 | Download | ----------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVED--GECYIQFF--SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRGAATKTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------- | |||||||||||||||||||
| 8 | 2rh1A | 0.32 | 0.30 | 0.89 | 4.02 | Download | ------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRAT-HQEAINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNKSRWYNQTPNRAKRVITTFRTGKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLCL------------------ | |||||||||||||||||||
| 9 | 3sn6R | 0.34 | 0.29 | 0.84 | 2.99 | Download | -------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAIN----CYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKI------------DKSEGRCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC------------------- | |||||||||||||||||||
| 10 | 3zpqA | 0.35 | 0.30 | 0.85 | 3.91 | Download | -------------GAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQA-LKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQS------------------RVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLLA------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
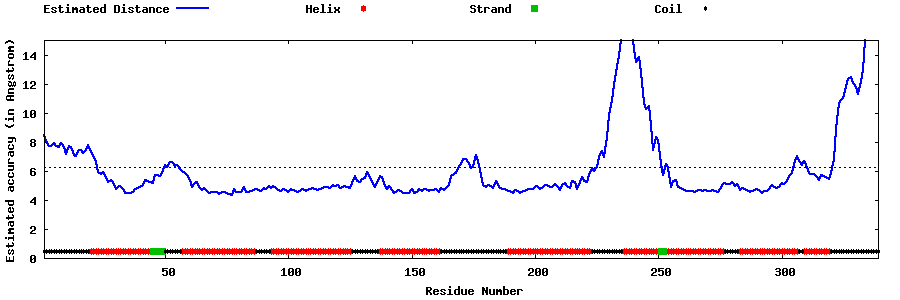
| Generated 3D models | Estimated local accuracy of models | ||
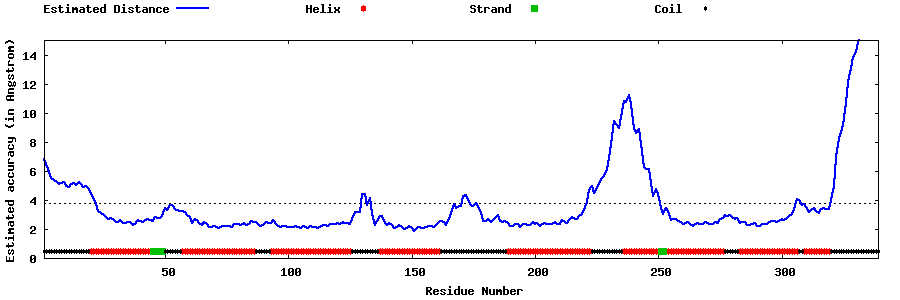 | |||
|
|
|||
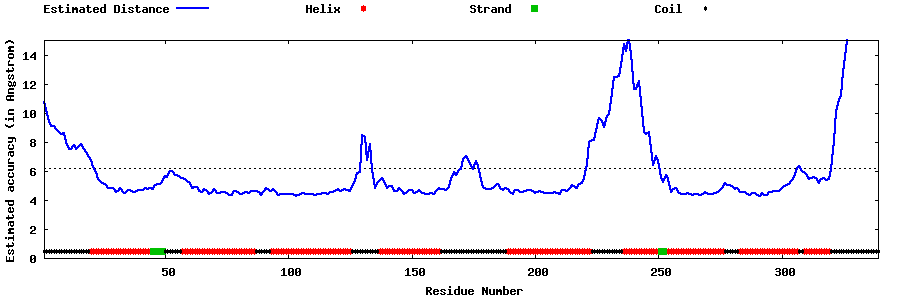 | |||
|
| |||
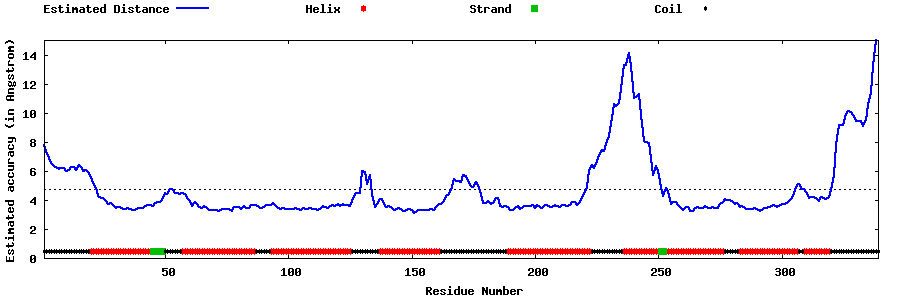 | |||
|
| |||
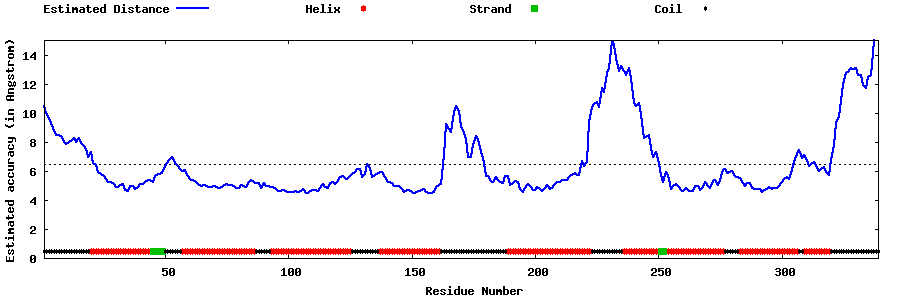 | |||
|
| |||
 | |||
|
| |||