

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MSAGNGTPWDATWNITVQWLAVDIACRTLMFLKLMATYSAAFLPVVIGLDRQAAVLNPLGSRSGVRKLLGAAWGLSFLLAFPQLFLFHTVHCAGPVPFTQCVTKGSFKAQWQETTYNLFTFCCLFLLPLTAMAICYSRIVLSVSRPQTRKGSHAPAGEFALPRSFDNCPRVRLRALRLALLILLTFILCWTPYYLLGMWYWFSPTMLTEVPPSLSHILFLLGLLNAPLDPLLYGAFTLGCRRGHQELSIDSSKEGSGRMLQEEIHAFRQLEVQKTVTSRRAGETKGISITSI | |
| CSSSSSHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9898614799999993814271789979999999999999999999999869985271838475788750499999999999999957788258875058985499860789999999999999999999999999999999977414666766555533211010578878799999999999999997999999999997565475525999999999999999899999998399999999998566667877655556777767666367777788888788536889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MSAGNGTPWDATWNITVQWLAVDIACRTLMFLKLMATYSAAFLPVVIGLDRQAAVLNPLGSRSGVRKLLGAAWGLSFLLAFPQLFLFHTVHCAGPVPFTQCVTKGSFKAQWQETTYNLFTFCCLFLLPLTAMAICYSRIVLSVSRPQTRKGSHAPAGEFALPRSFDNCPRVRLRALRLALLILLTFILCWTPYYLLGMWYWFSPTMLTEVPPSLSHILFLLGLLNAPLDPLLYGAFTLGCRRGHQELSIDSSKEGSGRMLQEEIHAFRQLEVQKTVTSRRAGETKGISITSI | |
| 4301000312001102332101200210130011000000000000000331000000021333020000000100000021000000024266443211000213233510320000000232233102102203120001023334445454454543344445423302100000000000010113131000000100132233402200100011100210021000000004301410240022234544644454444444444445444444445444343446 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CSSSSSHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MSAGNGTPWDATWNITVQWLAVDIACRTLMFLKLMATYSAAFLPVVIGLDRQAAVLNPLGSRSGVRKLLGAAWGLSFLLAFPQLFLFHTVHCAGPVPFTQCVTKGSFKAQWQETTYNLFTFCCLFLLPLTAMAICYSRIVLSVSRPQTRKGSHAPAGEFALPRSFDNCPRVRLRALRLALLILLTFILCWTPYYLLGMWYWFSPTMLTEVPPSLSHILFLLGLLNAPLDPLLYGAFTLGCRRGHQELSIDSSKEGSGRMLQEEIHAFRQLEVQKTVTSRRAGETKGISITSI | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.18 | 0.18 | 0.81 | 2.59 | Download | ALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.18 | 0.18 | 0.80 | 3.09 | Download | ALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKP----------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.18 | 0.18 | 0.81 | 3.08 | Download | ALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.17 | 0.19 | 0.96 | 1.56 | Download | TFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKRFIMIIFVWLWSVLWAIGPIFGWGAYTL--EGVLCNCSFDYISRD-STTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE--WVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKEDDKDAETEGESSDAAPSADAAQMKE------ | |||||||||||||||||||
| 5 | 4rnb | 0.18 | 0.20 | 0.85 | 1.17 | Download | LVTITCLPATLVVDITETWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPSTA-KRARNSIVIIWIVSCIIMIPQAIVMECSTVFKTTL--FTVCDERWGGEIYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSFWNQGSRSDLMSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMFARETVYAWFTFSHWLVYANSAANPIIYNFLSGKFREEFKAAFSC------------------------------------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.18 | 0.18 | 0.81 | 2.82 | Download | ALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVR-------------LLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.19 | 0.19 | 0.83 | 1.75 | Download | -----GFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHMIIFVWLWSVLWAIGPIFGWGAYTLEG--VLCNCSFDYI-SRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE--WVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWV----------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.19 | 0.19 | 0.77 | 3.18 | Download | LL---TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPT--SSKAQAVNVAIWALASVVGVPVAIMGSAQ---VEDEEIECLVEIPTPQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGS-------------REKDRNLRRITRLVLVVVAVFVGCWTPVQVFVLAQGLGVQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR----------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.18 | 0.18 | 0.81 | 2.86 | Download | ALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGA---VVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.18 | 0.18 | 0.81 | 3.28 | Download | ALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS-------------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
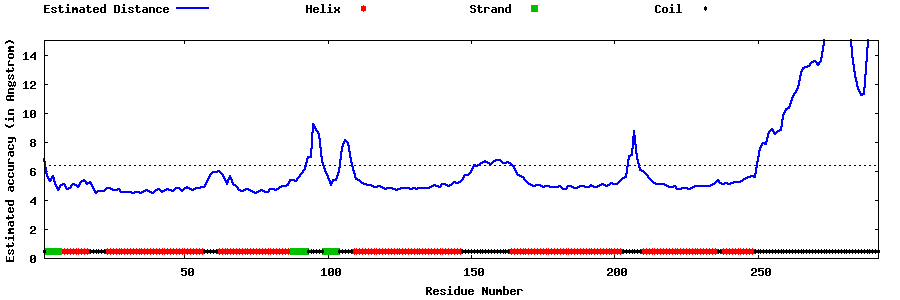
| Generated 3D models | Estimated local accuracy of models | ||
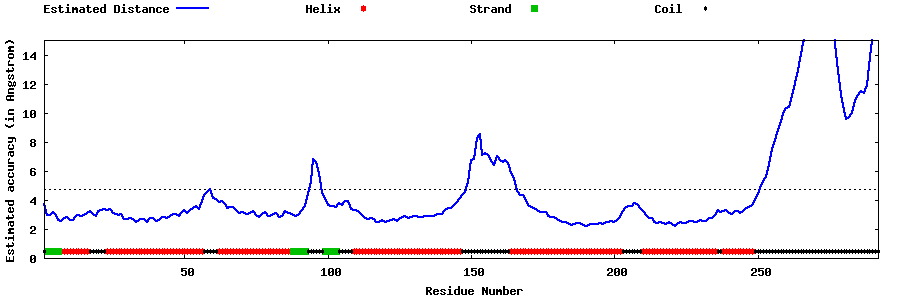 | |||
|
|
|||
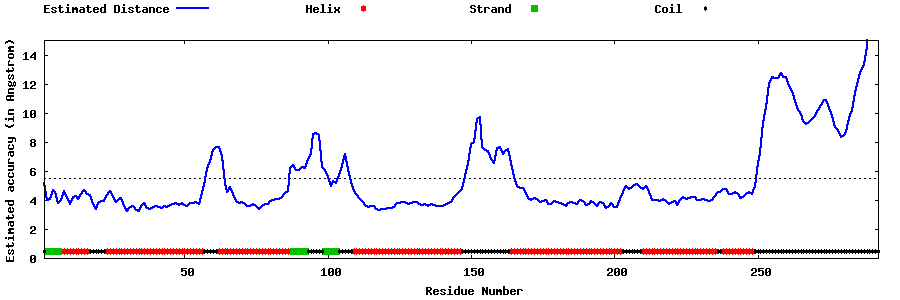 | |||
|
| |||
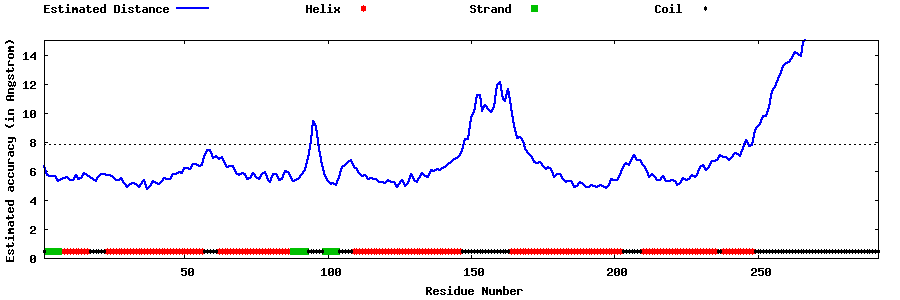 | |||
|
| |||
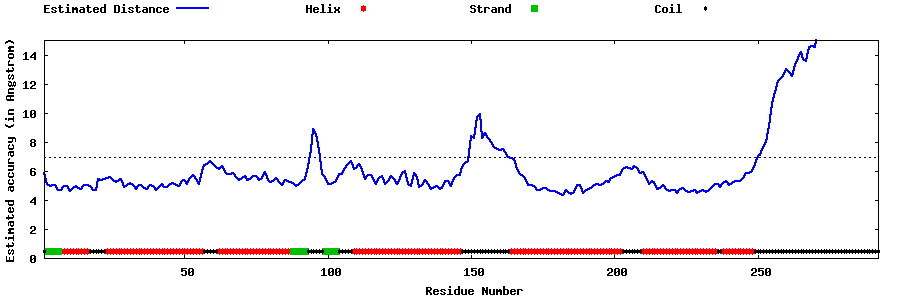 | |||
|
| |||
 | |||
|
| |||