

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MGPGEALLAGLLVMVLAVALLSNALVLLCCAYSAELRTRASGVLLVNLSLGHLLLAALDMPFTLLGVMRGRTPSAPGACQVIGFLDTFLASNAALSVAALSADQWLAVGFPLRYAGRLRPRYAGLLLGCAWGQSLAFSGAALGCSWLGYSSAFASCSLRLPPEPERPRFAAFTATLHAVGFVLPLAVLCLTSLQVHRVARRHCQRMDTVTMKALALLADLHPSVRQRCLIQQKRRRHRATRKIGIAIATFLICFAPYVMTRLAELVPFVTVNAQWGILSKCLTYSKAVADPFTYSLLRRPFRQVLAGMVHRLLKRTPRPASTHDSSLDVAGMVHQLLKRTPRPASTHNGSVDTENDSCLQQTH | |
| CCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 987999999999999999999999860102422877887757998999999999999989999999996833286478759999999999999999999999707513644768880878999885479999999999999988231026888538997579864300269999999999999999999999999999999988874110100013456786400246889999999999999999999999999999999999947887788999999999999998878998708999999999996274378999887768776667776644357899988888988899974102379 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MGPGEALLAGLLVMVLAVALLSNALVLLCCAYSAELRTRASGVLLVNLSLGHLLLAALDMPFTLLGVMRGRTPSAPGACQVIGFLDTFLASNAALSVAALSADQWLAVGFPLRYAGRLRPRYAGLLLGCAWGQSLAFSGAALGCSWLGYSSAFASCSLRLPPEPERPRFAAFTATLHAVGFVLPLAVLCLTSLQVHRVARRHCQRMDTVTMKALALLADLHPSVRQRCLIQQKRRRHRATRKIGIAIATFLICFAPYVMTRLAELVPFVTVNAQWGILSKCLTYSKAVADPFTYSLLRRPFRQVLAGMVHRLLKRTPRPASTHDSSLDVAGMVHQLLKRTPRPASTHNGSVDTENDSCLQQTH | |
| 632220000100120132133233000000011340313000000000010010001011102001102310000210020000000200100130200000300000010031343023200000000001203210120010001334442220102034314221210000100231132001000200120010013103414434444444446344444444334433332000000000001110211310000000001413021001000113032301210000000164003001300221143344434444343444433443344464444343343437444335448 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSSCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MGPGEALLAGLLVMVLAVALLSNALVLLCCAYSAELRTRASGVLLVNLSLGHLLLAALDMPFTLLGVMRGRTPSAPGACQVIGFLDTFLASNAALSVAALSADQWLAVGFPLRYAGRLRPRYAGLLLGCAWGQSLAFSGAALGCSWLGYSSAFASCSLRLPPEPERPRFAAFTATLHAVGFVLPLAVLCLTSLQVHRVARRHCQRMDTVTMKALALLADLHPSVRQRCLIQQKRRRHRATRKIGIAIATFLICFAPYVMTRLAELVPFVTVNAQWGILSKCLTYSKAVADPFTYSLLRRPFRQVLAGMVHRLLKRTPRPASTHDSSLDVAGMVHQLLKRTPRPASTHNGSVDTENDSCLQQTH | |||||||||||||||||||||||||
| 1 | 3sn6R | 0.17 | 0.18 | 0.78 | 2.63 | Download | -EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTV-TNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCY---AEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR--------------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC----------------------------------------------------- | |||||||||||||||||||
| 2 | 4amjA | 0.19 | 0.19 | 0.80 | 4.18 | Download | --QWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQT-LTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQ--------------IRKIDRASKRKTSRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLL------------------------------------------------------ | |||||||||||||||||||
| 3 | 3sn6R | 0.17 | 0.18 | 0.78 | 3.43 | Download | -EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQT-VTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQE---AINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSE--------------GRC------LKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC----------------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.15 | 0.18 | 0.94 | 1.55 | Download | PDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQ-TPANMFIINLAFSDFTFSLVNFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPI-FGWGAYTGVLCNCSFDYISRDST--TRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLN--------AKELRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKD----A-----ETEIP--AGESSDAAPSADAAQMKE | |||||||||||||||||||
| 5 | 3d4s | 0.15 | 0.19 | 0.84 | 1.20 | Download | ---WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQT-VTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHEAINCYAEE-TCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDKAGRNTNGVISTGTWDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL---------------------------------------------------- | |||||||||||||||||||
| 6 | 3sn6R | 0.17 | 0.18 | 0.78 | 3.05 | Download | --VWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQT-VTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCYA---EETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR--------------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC----------------------------------------------------- | |||||||||||||||||||
| 7 | 3d4s | 0.16 | 0.19 | 0.83 | 1.76 | Download | ----VVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQT-VTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHEAINCYAEETCC-DFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDEGGRNTNGVISTGTWDAYKFCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLC----------------------------------------------------- | |||||||||||||||||||
| 8 | 4djhA | 0.18 | 0.21 | 0.82 | 4.13 | Download | SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKT-ATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDECSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSKRVITTFRTGTWDAYREKD--------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALG--SAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------------------------------- | |||||||||||||||||||
| 9 | 3sn6R | 0.17 | 0.18 | 0.78 | 3.32 | Download | -EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQT-VTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCYAE---ETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR--------------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLC----------------------------------------------------- | |||||||||||||||||||
| 10 | 3zpqA | 0.20 | 0.18 | 0.78 | 3.56 | Download | LSQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTL-TNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQ--------------------------SRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLLA----------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
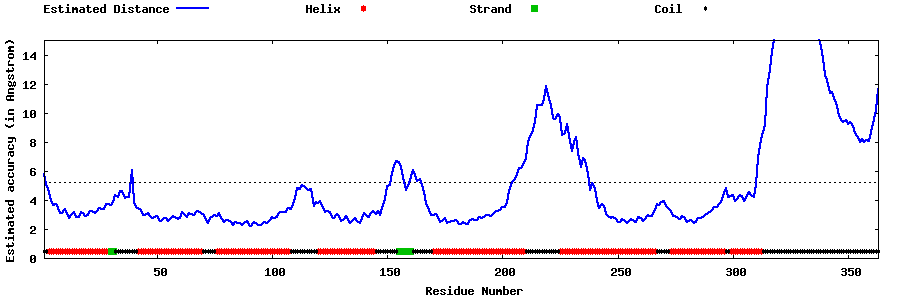
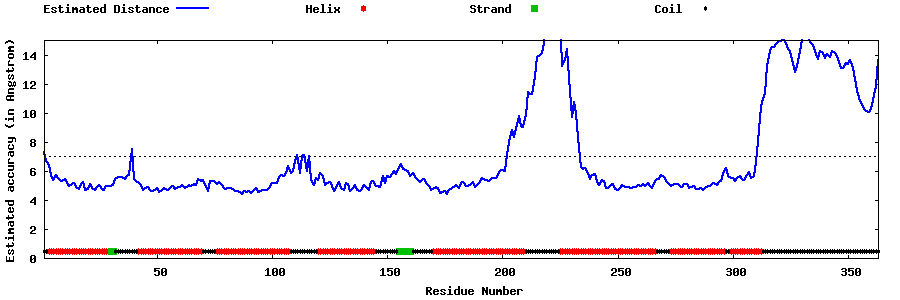
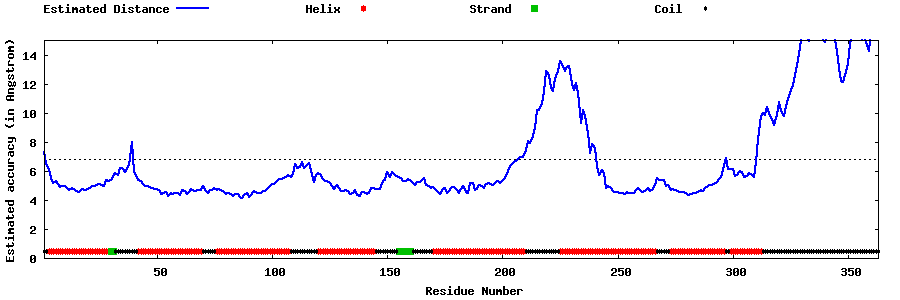
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||