

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNEPLDYLANASDFPDYAAAFGNCTDENIPLKMHYLPVIYGIIFLVGFPGNAVVISTYIFKMRPWKSSTIIMLNLACTDLLYLTSLPFLIHYYASGENWIFGDFMCKFIRFSFHFNLYSSILFLTCFSIFRYCVIIHPMSCFSIHKTRCAVVACAVVWIISLVAVIPMTFLITSTNRTNRSACLDLTSSDELNTIKWYNLILTATTFCLPLVIVTLCYTTIIHTLTHGLQTDSCLKQKARRLTILLLLAFYVCFLPFHILRVIRIESRLLSISCSIENQIHEAYIVSRPLAALNTFGNLLLYVVVSDNFQQAVCSTVRCKVSGNLEQAKKISYSNNP | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCC | |
| 9999778767778988766778777665217784699999999999998899989987750677869999999999999999995899999996699877886889999999999999999999999997613201003001255620366778999999999998999951014679963896159941258999999999999999999999999999999975510257643441889999999999998569999999999987558990699999999999999999999787998983288899999998352045873325687789997 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNEPLDYLANASDFPDYAAAFGNCTDENIPLKMHYLPVIYGIIFLVGFPGNAVVISTYIFKMRPWKSSTIIMLNLACTDLLYLTSLPFLIHYYASGENWIFGDFMCKFIRFSFHFNLYSSILFLTCFSIFRYCVIIHPMSCFSIHKTRCAVVACAVVWIISLVAVIPMTFLITSTNRTNRSACLDLTSSDELNTIKWYNLILTATTFCLPLVIVTLCYTTIIHTLTHGLQTDSCLKQKARRLTILLLLAFYVCFLPFHILRVIRIESRLLSISCSIENQIHEAYIVSRPLAALNTFGNLLLYVVVSDNFQQAVCSTVRCKVSGNLEQAKKISYSNNP | |
| 6634433333313223244434314434340221000220120033033232200100012334331010000000100000000020100000344321001001100102113111100100000031001000000202322332100000000110000001010001203647322101010357313211011122123113312210120000001102323466444220000000000000100123101000010013023340412400200110010001300331020000004400410140032224442653444346767 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCC MNEPLDYLANASDFPDYAAAFGNCTDENIPLKMHYLPVIYGIIFLVGFPGNAVVISTYIFKMRPWKSSTIIMLNLACTDLLYLTSLPFLIHYYASGENWIFGDFMCKFIRFSFHFNLYSSILFLTCFSIFRYCVIIHPMSCFSIHKTRCAVVACAVVWIISLVAVIPMTFLITSTNRTNRSACLDLTSSDELNTIKWYNLILTATTFCLPLVIVTLCYTTIIHTLTHGLQTDSCLKQKARRLTILLLLAFYVCFLPFHILRVIRIESRLLSISCSIENQIHEAYIVSRPLAALNTFGNLLLYVVVSDNFQQAVCSTVRCKVSGNLEQAKKISYSNNP | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.23 | 0.21 | 0.87 | 3.41 | Download | ---------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLR-MKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.36 | 0.32 | 0.87 | 4.33 | Download | -------------------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGRKNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKEEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLTPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRL------------------------ | |||||||||||||||||||
| 3 | 4mbsA | 0.23 | 0.21 | 0.87 | 3.94 | Download | ---------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRM-KEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------- | |||||||||||||||||||
| 4 | 4djh | 0.24 | 0.25 | 0.83 | 1.54 | Download | ----------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS----------AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------ | |||||||||||||||||||
| 5 | 4xnv | 0.36 | 0.33 | 0.86 | 1.16 | Download | --------------------SFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRNKTITCYDTTSDEYLRSYFIYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRR------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.23 | 0.21 | 0.87 | 3.46 | Download | ----------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE-KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------- | |||||||||||||||||||
| 7 | 4djh | 0.24 | 0.25 | 0.82 | 1.68 | Download | -------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----A-----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.24 | 0.22 | 0.86 | 4.44 | Download | ----------------------PCQKINVQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPSQYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRE-----KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.23 | 0.21 | 0.87 | 3.54 | Download | ---------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYSYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRM-KEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------- | |||||||||||||||||||
| 10 | 4n6hA | 0.20 | 0.23 | 0.94 | 3.99 | Download | LEDKSPDSTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKERRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
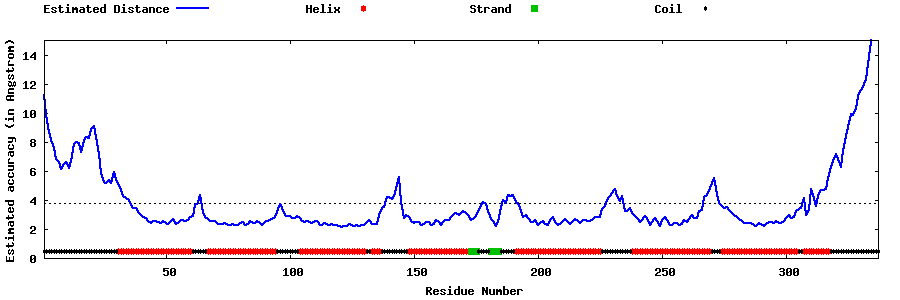 | |||
|
|
|||
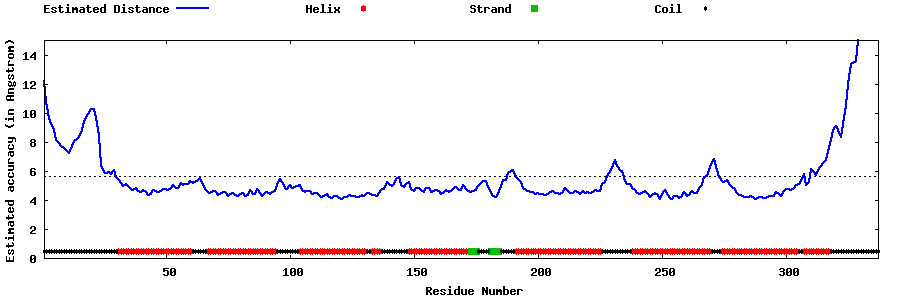 | |||
|
| |||
 | |||
|
| |||
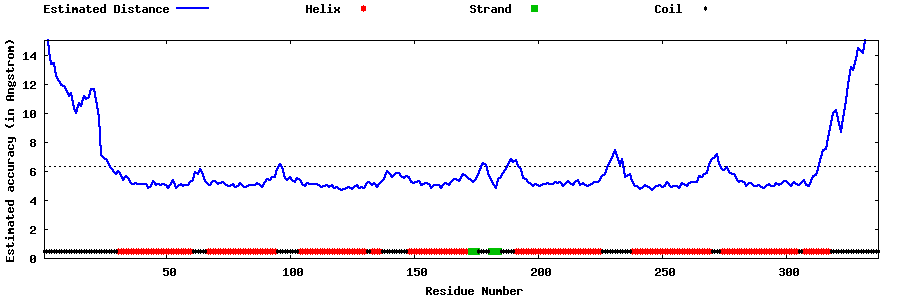 | |||
|
| |||
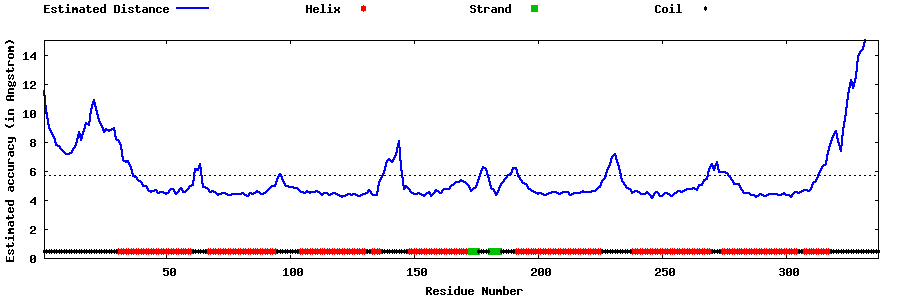 | |||
|
| |||