

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNNNTTCIQPSMISSMALPIIYILLCIVGVFGNTLSQWIFLTKIGKKTSTHIYLSHLVTANLLVCSAMPFMSIYFLKGFQWEYQSAQCRVVNFLGTLSMHASMFVSLLILSWIAISRYATLMQKDSSQETTSCYEKIFYGHLLKKFRQPNFARKLCIYIWGVVLGIIIPVTVYYSVIEATEGEESLCYNRQMELGAMISQIAGLIGTTFIGFSFLVVLTSYYSFVSHLRKIRTCTSIMEKDLTYSSVKRHLLVIQILLIVCFLPYSIFKPIFYVLHQRDNCQQLNYLIETKNILTCLASARSSTDPIIFLLLDKTFKKTLYNLFTKSNSAHMQSYG | |
| CCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCC | |
| 999988999601889989999999999999999999999884478885899999999999999999899999999569978768777787399998442711999999999999997899635062244444321100014665213055799989999999999998999986223537898789946798610379999999998866698999999999999999604678764212323003699999999999998688999999999971379945899999999999999999999999999941888999999995777898746699 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNNNTTCIQPSMISSMALPIIYILLCIVGVFGNTLSQWIFLTKIGKKTSTHIYLSHLVTANLLVCSAMPFMSIYFLKGFQWEYQSAQCRVVNFLGTLSMHASMFVSLLILSWIAISRYATLMQKDSSQETTSCYEKIFYGHLLKKFRQPNFARKLCIYIWGVVLGIIIPVTVYYSVIEATEGEESLCYNRQMELGAMISQIAGLIGTTFIGFSFLVVLTSYYSFVSHLRKIRTCTSIMEKDLTYSSVKRHLLVIQILLIVCFLPYSIFKPIFYVLHQRDNCQQLNYLIETKNILTCLASARSSTDPIIFLLLDKTFKKTLYNLFTKSNSAHMQSYG | |
| 753424043444011000022012002313323220010001224432001000100020221100001010000035340100100000000023321110001001100100310010001002023334333333333333312122001000010021012001010001203425743200000222644220010012213311233221001000000110131344444456444211000000000000100122101010011003225404233002001110100013000210000000046014101400333344436658 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCC MNNNTTCIQPSMISSMALPIIYILLCIVGVFGNTLSQWIFLTKIGKKTSTHIYLSHLVTANLLVCSAMPFMSIYFLKGFQWEYQSAQCRVVNFLGTLSMHASMFVSLLILSWIAISRYATLMQKDSSQETTSCYEKIFYGHLLKKFRQPNFARKLCIYIWGVVLGIIIPVTVYYSVIEATEGEESLCYNRQMELGAMISQIAGLIGTTFIGFSFLVVLTSYYSFVSHLRKIRTCTSIMEKDLTYSSVKRHLLVIQILLIVCFLPYSIFKPIFYVLHQRDNCQQLNYLIETKNILTCLASARSSTDPIIFLLLDKTFKKTLYNLFTKSNSAHMQSYG | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.19 | 0.22 | 0.88 | 3.14 | Download | --------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEA-KRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILP----SLILLNMYASILLLATISADRFLLVFKPAWC----------------QRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVLCGVDHDK--RRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSA--------RETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFL--EPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR- | |||||||||||||||||||
| 2 | 4xnwA | 0.19 | 0.21 | 0.87 | 4.48 | Download | -SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKTDWIFGDAMCKLQRFI----FHVNLYGSILFLTCISAHRYSGVVYPLKS----------------LGRLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKT-ITCYDTTSDEYLRSYFYSMCTTVAMFCVPLVLILGCYGLIVRALIY--------KEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLPAMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR-------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.23 | 0.25 | 0.90 | 3.44 | Download | --SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGE----LLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----------------LDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG--AVVCMLQFPSPSWYWDTVTKICVFLFFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV---DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------- | |||||||||||||||||||
| 4 | 4djh | 0.22 | 0.24 | 0.85 | 1.53 | Download | ----------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYL-MNSWPFGDVLCKIVLSIDYY----NMFTSIFTLTMMSVDRYIAVCHPVK----------------ALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYWWDLFMICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------- | |||||||||||||||||||
| 5 | 4djh | 0.22 | 0.24 | 0.85 | 1.17 | Download | ----------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDY----YNMFTSIFTLTMMSVDRYIAVCHPVKAL----------------DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDDYSWWDLKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------- | |||||||||||||||||||
| 6 | 4n6hA | 0.23 | 0.25 | 0.89 | 3.32 | Download | ----GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYY----NMFTSIFTLTMMSVDRYIAVCHPV----------------KALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD--GAVVCMLQFPPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRR---DPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------- | |||||||||||||||||||
| 7 | 4djh | 0.21 | 0.24 | 0.39 | 1.71 | Download | -----------PAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVY-LMNSWPFGDVLCKIVLSIDYYN----MFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLS--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.21 | 0.23 | 0.85 | 4.08 | Download | -----PCQKINQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGF----FSGIFFIILLTIDRYLAVVH----------------AVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGL--HYTCSSHFPYSQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREK----------KRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ----------- | |||||||||||||||||||
| 9 | 5o9hA | 0.19 | 0.22 | 0.88 | 3.32 | Download | --------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFE-AKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILP----SLILLNMYASILLLATISADRFLLVFKPAWCQRFR----------------GAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVLCGVDHDK--RRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWS--------ARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFL--EPSSPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR- | |||||||||||||||||||
| 10 | 4n6hA | 0.22 | 0.23 | 0.90 | 3.77 | Download | LGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFG----ELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALD----------------FRTPAKAKLINICIWVLASGVGVPIMVMAVTRP--RDGAVVCMLQFPSPSWYWDTVTKICVFLFFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRR---DPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
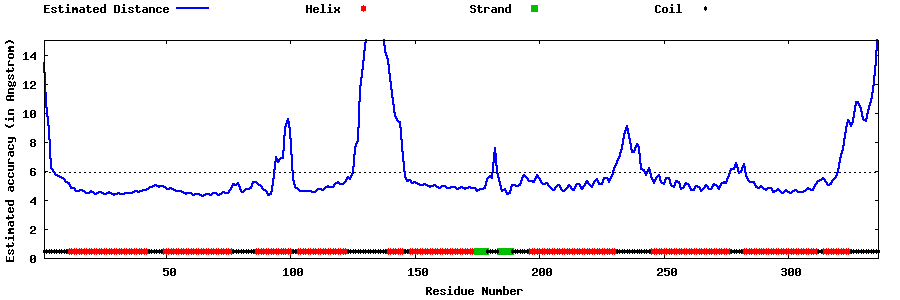
| Generated 3D models | Estimated local accuracy of models | ||
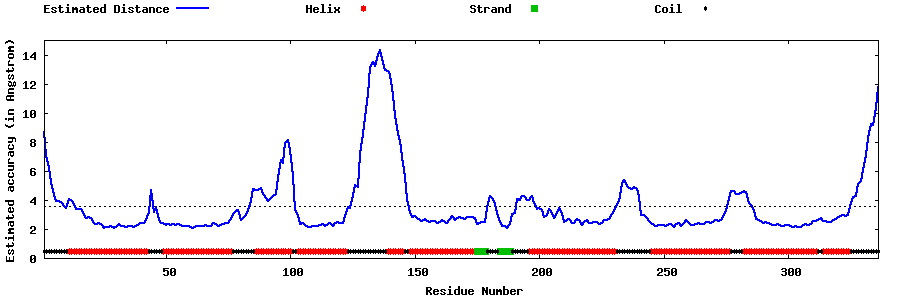 | |||
|
|
|||
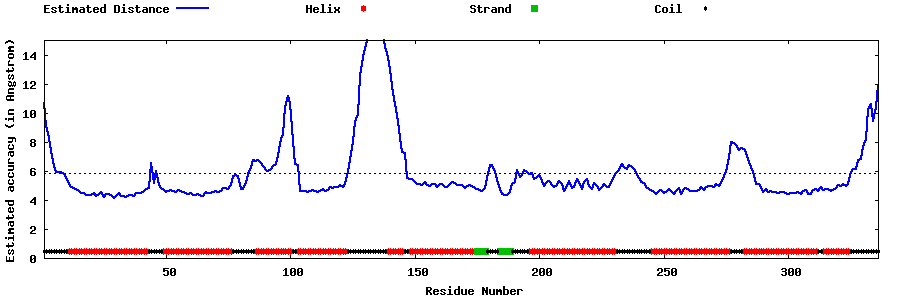 | |||
|
| |||
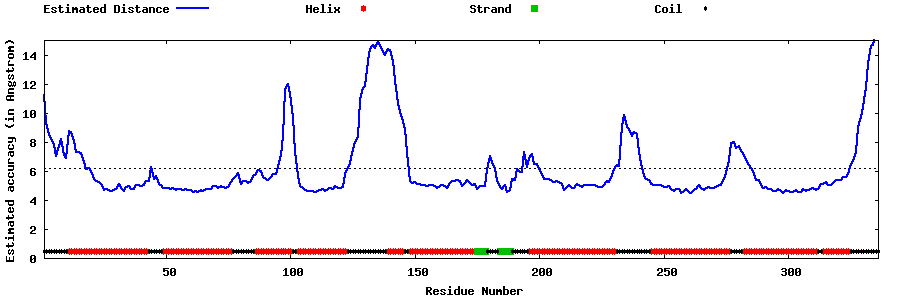 | |||
|
| |||
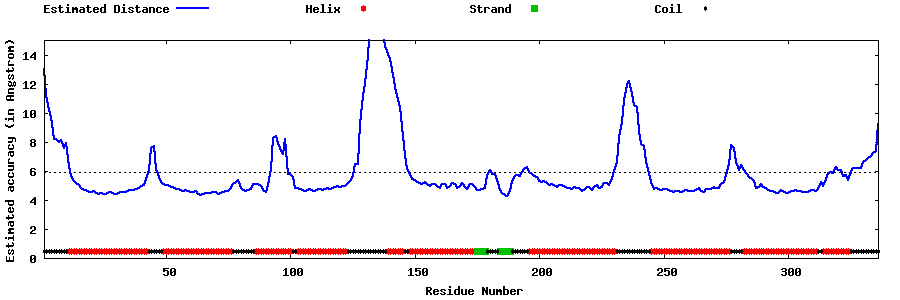 | |||
|
| |||
 | |||
|
| |||