

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDPTISTLDTELTPINGTEETLCYKQTLSLTVLTCIVSLVGLTGNAVVLWLLGCRMRRNAFSIYILNLAAADFLFLSGRLIYSLLSFISIPHTISKILYPVMMFSYFAGLSFLSAVSTERCLSVLWPIWYRCHRPTHLSAVVCVLLWALSLLRSILEWMLCGFLFSGADSAWCQTSDFITVAWLIFLCVVLCGSSLVLLIRILCGSRKIPLTRLYVTILLTVLVFLLCGLPFGIQFFLFLWIHVDREVLFCHVHLVSIFLSALNSSANPIIYFFVGSFRQRQNRQNLKLVLQRALQDASEVDEGGGQLPEEILELSGSRLEQ | |
| CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHSSSHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9999888888889999987554411799999999999999998899989987760689785999999999999999989999985357766569999999999999999999999999988999988760568874158999999999999998479887211358998315589999999999999999889999999999862577778842441220205779986799999999999867812799999999999999999786696625478788999999999976454588888999998889876467300049 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDPTISTLDTELTPINGTEETLCYKQTLSLTVLTCIVSLVGLTGNAVVLWLLGCRMRRNAFSIYILNLAAADFLFLSGRLIYSLLSFISIPHTISKILYPVMMFSYFAGLSFLSAVSTERCLSVLWPIWYRCHRPTHLSAVVCVLLWALSLLRSILEWMLCGFLFSGADSAWCQTSDFITVAWLIFLCVVLCGSSLVLLIRILCGSRKIPLTRLYVTILLTVLVFLLCGLPFGIQFFLFLWIHVDREVLFCHVHLVSIFLSALNSSANPIIYFFVGSFRQRQNRQNLKLVLQRALQDASEVDEGGGQLPEEILELSGSRLEQ | |
| 8334434443433334334532432210101321321033333330100000202343101000000200010010100000102310210101012013323311210010000012123000000010232133210000000001000000111000000113664220010011123131333221212100000011023346344300000000002001302332222100100031433200200121010001300021000000003601520241022003300443344566654344444434446368 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHSSSHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDPTISTLDTELTPINGTEETLCYKQTLSLTVLTCIVSLVGLTGNAVVLWLLGCRMRRNAFSIYILNLAAADFLFLSGRLIYSLLSFISIPHTISKILYPVMMFSYFAGLSFLSAVSTERCLSVLWPIWYRCHRPTHLSAVVCVLLWALSLLRSILEWMLCGFLFSGADSAWCQTSDFITVAWLIFLCVVLCGSSLVLLIRILCGSRKIPLTRLYVTILLTVLVFLLCGLPFGIQFFLFLWIHVDREVLFCHVHLVSIFLSALNSSANPIIYFFVGSFRQRQNRQNLKLVLQRALQDASEVDEGGGQLPEEILELSGSRLEQ | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.21 | 0.23 | 0.86 | 3.13 | Download | --------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYKVLCGVDHDKRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLESPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTE---ESVVR--------------------- | |||||||||||||||||||
| 2 | 4ntjA | 0.18 | 0.21 | 0.77 | 3.94 | Download | ------------------------ITQVLFPLLYTVLFFVGLITNGLAMRIFFQIRSKSNFIIFLKNTVISDLLMILTFPFKILSDAKLLRTFVCQVTSVIFYFTMYISISFLGLITIDRYQKTTRPF---KTKNLLGAKILSVVIWAFMFLLSLPNMIL-----KSEFGLVWHEIVNYICQVIFWINFLIVIVCYTLITKELYRSR----KKVNVKVFIIIAVFFICFVPFHFARIPYTLSDCTAENTLFYVKESTLWLTSLNACLNPFIYFFLCKSFRNSLIS------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.23 | 0.86 | 3.66 | Download | --------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKKTATNIYIFNLALADALATSTLPFQSAKYLWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------ | |||||||||||||||||||
| 4 | 4ib4 | 0.14 | 0.21 | 0.84 | 1.51 | Download | --------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLKKLQYATNYFLMSLAVADLLVGLFVMPIALLAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETN--PNNITCVLFLFGSLAAFFTPLAIMIVTYFLTIHALQKKAAISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFN----KTFRDAFGRYITCNYR-------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.24 | 0.23 | 0.81 | 1.15 | Download | ----------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYKMKTATNIYIFNLALADALVTTTMPFQSTMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.21 | 0.23 | 0.86 | 3.09 | Download | ---------------------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYKVLCGVDHDKRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLESPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTE---ESVVR--------------------- | |||||||||||||||||||
| 7 | 4ib4 | 0.14 | 0.21 | 0.81 | 1.69 | Download | -------------------------KLHWAALLILMVIIPTIGGNTLVILAVSLKKLQYATNYFLMSLAVADLLVGLFVMPIALLAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIGIETN-P--NNITCVLTKERFSFFTPLAIMIVTYFLTIHALQKKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITC--------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.21 | 0.23 | 0.84 | 3.76 | Download | ---------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRKSMTDIYLLNLAISDLFFLLTVPFWAHYAQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREKKRHRD--VRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGSNRLDQAMQVTETLGMTHCCINPIIYAFVG----EEFRNYLLVFFQ------------------------------ | |||||||||||||||||||
| 9 | 5o9hA | 0.20 | 0.23 | 0.86 | 3.27 | Download | --------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLESPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTE---ESVVR--------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.23 | 0.22 | 0.84 | 4.09 | Download | -----------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKKTATNIYIFNLALADALATSTLPFQSVNYTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
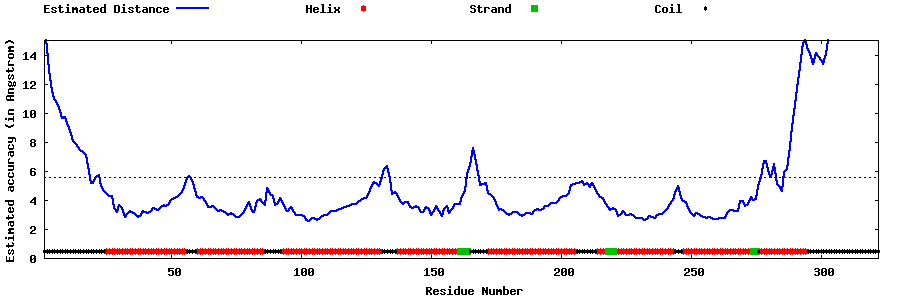 | |||
|
|
|||
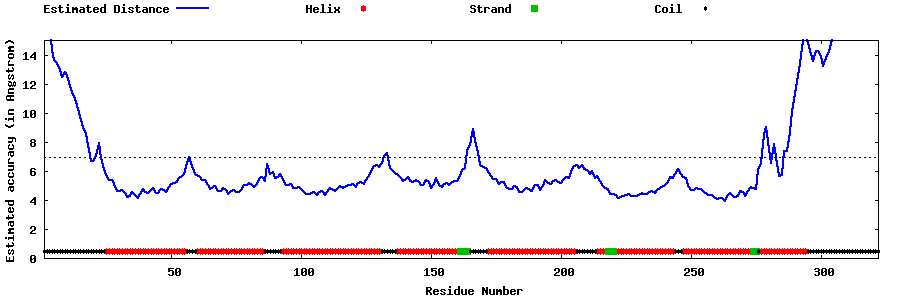 | |||
|
| |||
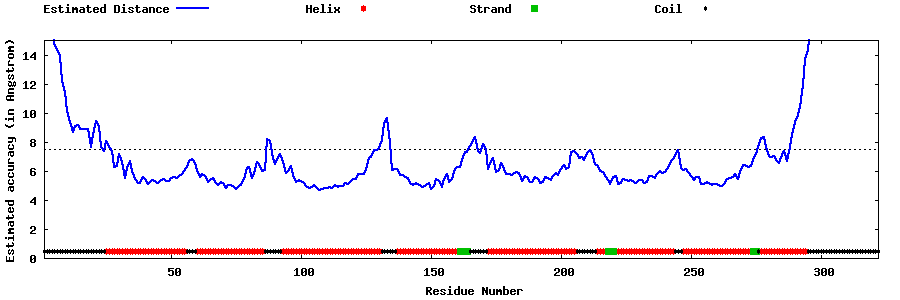 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||