

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDPTVPVFGTKLTPINGREETPCYNQTLSFTVLTCIISLVGLTGNAVVLWLLGYRMRRNAVSIYILNLAAADFLFLSFQIIRLPLRLINISHLIRKILVSVMTFPYFTGLSMLSAISTERCLSVLWPIWYRCRRPTHLSAVVCVLLWGLSLLFSMLEWRFCDFLFSGADSSWCETSDFIPVAWLIFLCVVLCVSSLVLLVRILCGSRKMPLTRLYVTILLTVLVFLLCGLPFGILGALIYRMHLNLEVLYCHVYLVCMSLSSLNSSANPIIYFFVGSFRQRQNRQNLKLVLQRALQDKPEVDKGEGQLPEESLELSGSRLGP | |
| CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHSSSSSHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9999888888889999887887622899999999999999998899989988753689775999999999999999999999999984566679999999999999999999999999978999987662467862489999999999999999689988233348998440158999999999999999999999999999861687778750040443024649986799999999998536840899999999999999999787898862388889999999999863244688888999997868883037788896 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MDPTVPVFGTKLTPINGREETPCYNQTLSFTVLTCIISLVGLTGNAVVLWLLGYRMRRNAVSIYILNLAAADFLFLSFQIIRLPLRLINISHLIRKILVSVMTFPYFTGLSMLSAISTERCLSVLWPIWYRCRRPTHLSAVVCVLLWGLSLLFSMLEWRFCDFLFSGADSSWCETSDFIPVAWLIFLCVVLCVSSLVLLVRILCGSRKMPLTRLYVTILLTVLVFLLCGLPFGILGALIYRMHLNLEVLYCHVYLVCMSLSSLNSSANPIIYFFVGSFRQRQNRQNLKLVLQRALQDKPEVDKGEGQLPEESLELSGSRLGP | |
| 8334344342433334336423232210101221220022133331100000202343101000000001000000101013111320130100012013213311210020000002000000000010233133210000000002000100100000000213664220100010223133333221212100000011124347344300000000001000001222211100100131323310100121010001300021000000003401520240022003310434344566654334443434446468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHSSSSSHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDPTVPVFGTKLTPINGREETPCYNQTLSFTVLTCIISLVGLTGNAVVLWLLGYRMRRNAVSIYILNLAAADFLFLSFQIIRLPLRLINISHLIRKILVSVMTFPYFTGLSMLSAISTERCLSVLWPIWYRCRRPTHLSAVVCVLLWGLSLLFSMLEWRFCDFLFSGADSSWCETSDFIPVAWLIFLCVVLCVSSLVLLVRILCGSRKMPLTRLYVTILLTVLVFLLCGLPFGILGALIYRMHLNLEVLYCHVYLVCMSLSSLNSSANPIIYFFVGSFRQRQNRQNLKLVLQRALQDKPEVDKGEGQLPEESLELSGSRLGP | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.22 | 0.23 | 0.86 | 3.10 | Download | --------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIV-PFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREYFPPKVLCGVVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLESPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTE------ESVVR------------------ | |||||||||||||||||||
| 2 | 4ntjA | 0.17 | 0.21 | 0.79 | 3.94 | Download | -------------------TRDYKITQVLFPLLYTVLFFVGLITNGLAMRIFFQIRSKSNFIIFLKNTVISDLLMILTFPFKILSDAKLLRTFVCQVTSVIFYFTMYISISFLGLITIDRYQKTTRPF---KTKNLLGAKILSVVIWAFMFLLSLPNMIL-----KSEFGLVWHEIVNYICQVIFWINFLIVIVCYTLITKELYRSR----KKVNVKVFIIIAVFFICFVPFHFARIPYTLSQTRAENTLFYVKESTLWLTSLNACLNPFIYFFLCKSFRNSLIS------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.25 | 0.86 | 3.58 | Download | --------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLMPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------ | |||||||||||||||||||
| 4 | 4ib4 | 0.14 | 0.21 | 0.84 | 1.51 | Download | --------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLKKLQYATNYFLMSLAVADLLVGLFVMPIALLTMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIET--NPNNITCVLFLFGSLAAFFTPLAIMIVTYFLTIHALQKKAAISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFN----KTFRDAFGRYITCNYR-------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.23 | 0.25 | 0.81 | 1.15 | Download | ----------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRTKMKTATNIYIFNLALADALVTTTMPFQSTVNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.22 | 0.23 | 0.86 | 3.15 | Download | ---------------------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEDHDKRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLESPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------ | |||||||||||||||||||
| 7 | 4ib4 | 0.13 | 0.21 | 0.81 | 1.71 | Download | --------------------------LHWAALLILMVIIPTIGGNTLVILAVLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNP--NNITCVLTKFLAAFFTPLAIMIVTYFLTIHALQKKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCN-------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.20 | 0.22 | 0.84 | 3.76 | Download | ---------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRKSMTDIYLLNLAISDLFFLLTVPFWAHYAAWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFCSSHFPYSQQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREKKRHRD--VRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGSNRLDQAMQVTETLGMTHCCINPIIYAFVG----EEFRNYLLVFFQ------------------------------ | |||||||||||||||||||
| 9 | 5o9hA | 0.21 | 0.23 | 0.86 | 3.30 | Download | --------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIV-PFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLESPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------------ | |||||||||||||||||||
| 10 | 5c1mA | 0.24 | 0.25 | 0.84 | 4.08 | Download | --------GSHSLPQTGS---PSMVTAITIMALYSIVCVVGLFGNFLVMYVIVYTKMKTATNIYIFNLALADALATSTLPFQSVNYTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
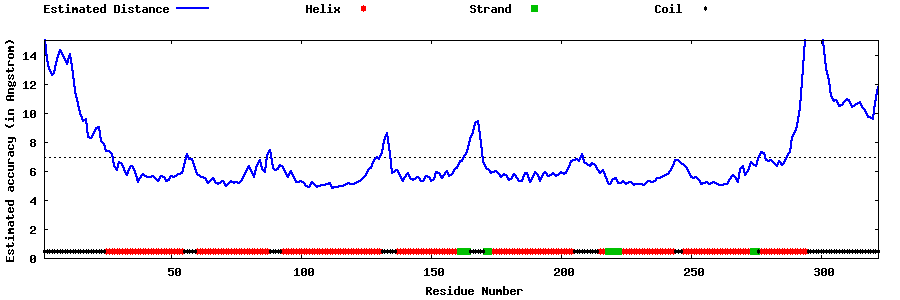
| Generated 3D models | Estimated local accuracy of models | ||
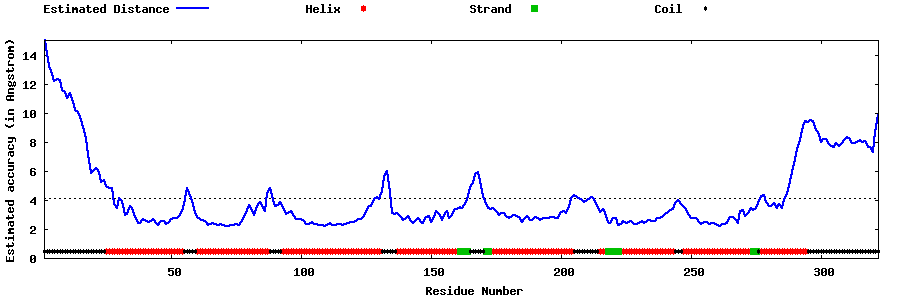 | |||
|
|
|||
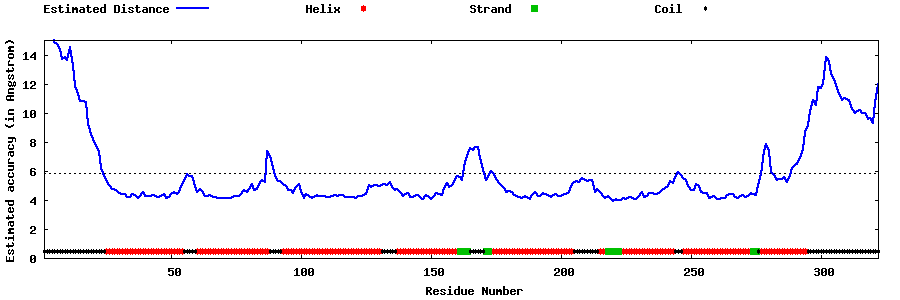 | |||
|
| |||
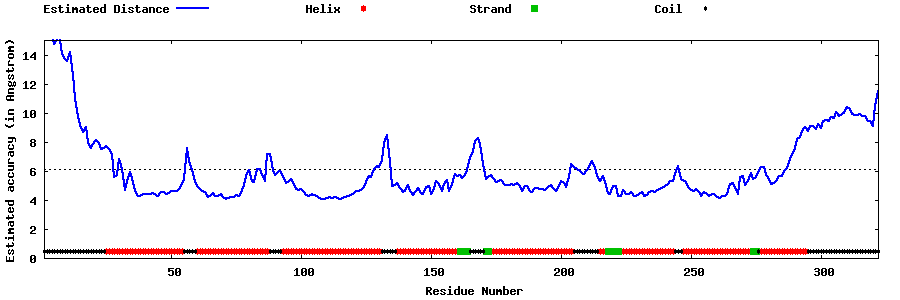 | |||
|
| |||
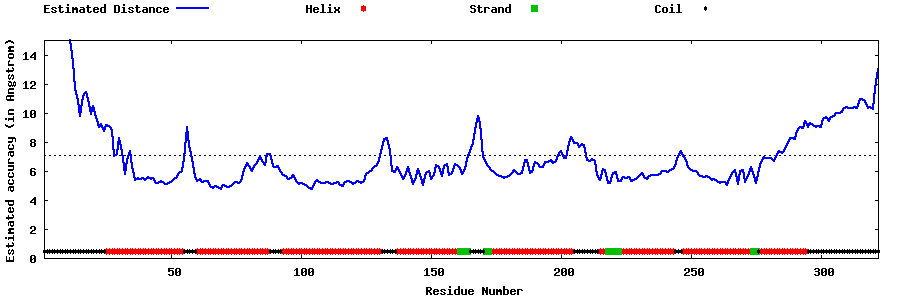 | |||
|
| |||
 | |||
|
| |||