

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MWSCSWFNGTGLVEELPACQDLQLGLSLLSLLGLVVGVPVGLCYNALLVLANLHSKASMTMPDVYFVNMAVAGLVLSALAPVHLLGPPSSRWALWSVGGEVHVALQIPFNVSSLVAMYSTALLSLDHYIERALPRTYMASVYNTRHVCGFVWGGALLTSFSSLLFYICSHVSTRALECAKMQNAEAADATLVFIGYVVPALATLYALVLLSRVRREDTPLDRDTGRLEPSAHRLLVATVCTQFGLWTPHYLILLGHTVIISRGKPVDAHYLGLLHFVKDFSKLLAFSSSFVTPLLYRYMNQSFPSKLQRLMKKLPCGDRHCSPDHMGVQQVLA | |
| CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCC | |
| 998888778999655555544321555287899999999988569975374750887696999999999999999999639999999972888770232589999999899999999999999998572040118400147678862129999999999699998436876897499707898779999987554999999999999999999965486531120100148888999999999998999999999999984588887689999999999999999999989999999819787799999997478878886888887415779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MWSCSWFNGTGLVEELPACQDLQLGLSLLSLLGLVVGVPVGLCYNALLVLANLHSKASMTMPDVYFVNMAVAGLVLSALAPVHLLGPPSSRWALWSVGGEVHVALQIPFNVSSLVAMYSTALLSLDHYIERALPRTYMASVYNTRHVCGFVWGGALLTSFSSLLFYICSHVSTRALECAKMQNAEAADATLVFIGYVVPALATLYALVLLSRVRREDTPLDRDTGRLEPSAHRLLVATVCTQFGLWTPHYLILLGHTVIISRGKPVDAHYLGLLHFVKDFSKLLAFSSSFVTPLLYRYMNQSFPSKLQRLMKKLPCGDRHCSPDHMGVQQVLA | |
| 716042212133344343264130111111121121002312311200010001033321000031133111311000000010000026332201000100010112133111100010000031020000101233223210100000001000000201000120343743210001222430120010120233122001200000011024344364454444200000000000000001220000000100231333343132330020011101000030013110000000431242013002402134442444444344337 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCC MWSCSWFNGTGLVEELPACQDLQLGLSLLSLLGLVVGVPVGLCYNALLVLANLHSKASMTMPDVYFVNMAVAGLVLSALAPVHLLGPPSSRWALWSVGGEVHVALQIPFNVSSLVAMYSTALLSLDHYIERALPRTYMASVYNTRHVCGFVWGGALLTSFSSLLFYICSHVSTRALECAKMQNAEAADATLVFIGYVVPALATLYALVLLSRVRREDTPLDRDTGRLEPSAHRLLVATVCTQFGLWTPHYLILLGHTVIISRGKPVDAHYLGLLHFVKDFSKLLAFSSSFVTPLLYRYMNQSFPSKLQRLMKKLPCGDRHCSPDHMGVQQVLA | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.17 | 0.22 | 0.88 | 3.28 | Download | --------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME---TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV------DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK-PCG---------------- | |||||||||||||||||||
| 2 | 4xnwA | 0.13 | 0.18 | 0.88 | 3.97 | Download | -------------SSFKCALTKTGFQFYYLPAVYILVFIIGFLGNSVAIWMFVFHMKPWSGISVYMFNLALADFLYVLTLPALIFYYFNKT--DWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYPLKSLLKKKNAICISVLVWLIVVVAISPILFYSGTGVRKNKTITCYDTTSDEYYSMCTTVAMFCVPLVLILGCYGLIVRALIYKMKKEEVEEPLRRKSIYLVIIVLTVFAVSYIPFHVMKTMNLRARLDFQTPMCAFNDRVYATYQVTRGLASLNSCVNPILYFLAGDTFRRRLSR------------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.16 | 0.22 | 0.88 | 3.64 | Download | --------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME---TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRD------PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------- | |||||||||||||||||||
| 4 | 4djh | 0.17 | 0.18 | 0.83 | 1.51 | Download | ---------------------SPAIPVIITAVYSVVFV-VGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM---NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA------------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------------------- | |||||||||||||||||||
| 5 | 4djh | 0.16 | 0.18 | 0.83 | 1.15 | Download | ----------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN---SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREVDVIECSLQFPDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALG---SA---------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.16 | 0.22 | 0.87 | 3.29 | Download | ----------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME---TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVLDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRR------DPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG----------------- | |||||||||||||||||||
| 7 | 4djh | 0.16 | 0.18 | 0.82 | 1.67 | Download | -------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM---NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS----A--------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF--------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.17 | 0.22 | 0.88 | 3.31 | Download | -------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLMET---WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV--DIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK-PCG---------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.16 | 0.21 | 0.86 | 3.29 | Download | ---------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ----WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALRTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE----KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGL-NNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ--------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.14 | 0.18 | 0.86 | 4.48 | Download | -----------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMGT---WPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF-------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
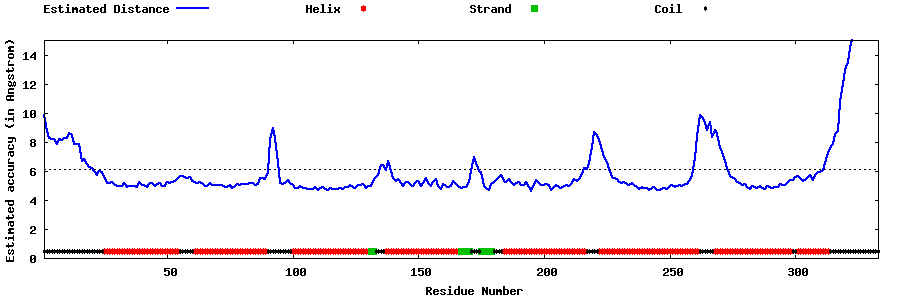
| Generated 3D models | Estimated local accuracy of models | ||
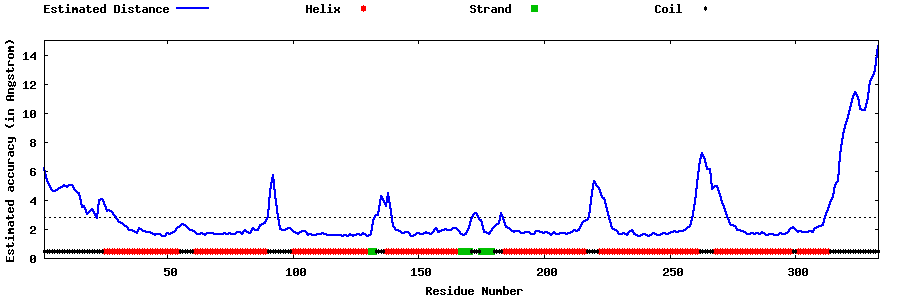 | |||
|
|
|||
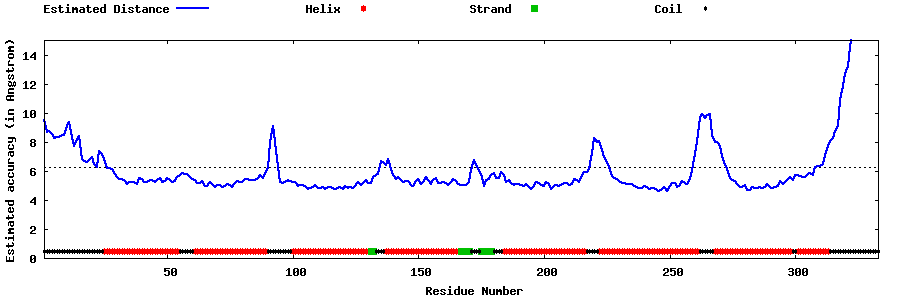 | |||
|
| |||
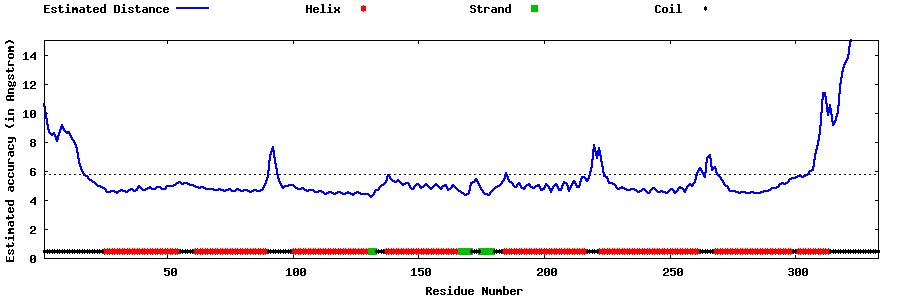 | |||
|
| |||
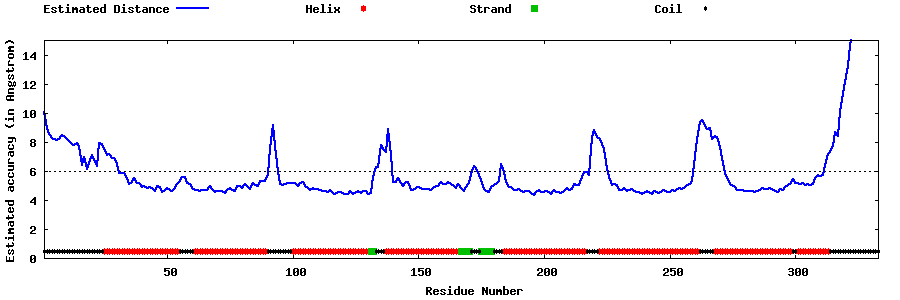 | |||
|
| |||
 | |||
|
| |||