

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MNPFHASCWNTSAELLNKSWNKEFAYQTASVVDTVILPSMIGIICSTGLVGNILIVFTIIRSRKKTVPDIYICNLAVADLVHIVGMPFLIHQWARGGEWVFGGPLCTIITSLDTCNQFACSAIMTVMSVDRYFALVQPFRLTRWRTRYKTIRINLGLWAASFILALPVWVYSKVIKFKDGVESCAFDLTSPDDVLWYTLYLTITTFFFPLPLILVCYILILCYTWEMYQQNKDARCCNPSVPKQRVMKLTKMVLVLVVVFILSAAPYHVIQLVNLQMEQPTLAFYVGYYLSICLSYASSSINPFLYILLSGNFQKRLPQIQRRATEKEINNMGNTLKSHF | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCC | |
| 9998999988877767888776556544212888008999999999998889986786434788748899999999999999998489999997699777986788699999999999999999999999899998535312163566788631799999999995999986348858995799860896267999999999999999999999999999999976357777632111145555556566778999999999979999999999956154389999999999999999989999999849999999999857646686677887577889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MNPFHASCWNTSAELLNKSWNKEFAYQTASVVDTVILPSMIGIICSTGLVGNILIVFTIIRSRKKTVPDIYICNLAVADLVHIVGMPFLIHQWARGGEWVFGGPLCTIITSLDTCNQFACSAIMTVMSVDRYFALVQPFRLTRWRTRYKTIRINLGLWAASFILALPVWVYSKVIKFKDGVESCAFDLTSPDDVLWYTLYLTITTFFFPLPLILVCYILILCYTWEMYQQNKDARCCNPSVPKQRVMKLTKMVLVLVVVFILSAAPYHVIQLVNLQMEQPTLAFYVGYYLSICLSYASSSINPFLYILLSGNFQKRLPQIQRRATEKEINNMGNTLKSHF | |
| 6443444322313422333334524343243010000022012002302321220010010133220000000000100000000000100000254421002001210100132011000000000021001000000202421323100000010110000001000001202537632210102024642220010131231133122000200000011013234444444444443444231000000000010010012210000001001333320010011001000130001102010000430141014003323644355445444644 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCC MNPFHASCWNTSAELLNKSWNKEFAYQTASVVDTVILPSMIGIICSTGLVGNILIVFTIIRSRKKTVPDIYICNLAVADLVHIVGMPFLIHQWARGGEWVFGGPLCTIITSLDTCNQFACSAIMTVMSVDRYFALVQPFRLTRWRTRYKTIRINLGLWAASFILALPVWVYSKVIKFKDGVESCAFDLTSPDDVLWYTLYLTITTFFFPLPLILVCYILILCYTWEMYQQNKDARCCNPSVPKQRVMKLTKMVLVLVVVFILSAAPYHVIQLVNLQMEQPTLAFYVGYYLSICLSYASSSINPFLYILLSGNFQKRLPQIQRRATEKEINNMGNTLKSHF | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.29 | 0.31 | 0.94 | 3.01 | Download | EAQAAAEQLKTTRNYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPR-DGAVVCMLQFPSPSWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.22 | 0.22 | 0.87 | 4.12 | Download | ----------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQK-EGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKKCGVGKDQFEEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.30 | 0.30 | 0.87 | 4.00 | Download | ---------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSK------EKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------- | |||||||||||||||||||
| 4 | 4djh | 0.31 | 0.32 | 0.85 | 1.52 | Download | -----------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKKTATNIYIFNLALADALVTTTMPFQST-VYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLFPDDDYSWWDLFMKVIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNILQTAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------- | |||||||||||||||||||
| 5 | 4djh | 0.31 | 0.32 | 0.85 | 1.16 | Download | -----------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIKTPAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.29 | 0.30 | 0.87 | 3.48 | Download | ---------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKKTATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG-AVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRL------LSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------- | |||||||||||||||||||
| 7 | 4djh | 0.31 | 0.32 | 0.84 | 1.69 | Download | -------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTMKTATNIYIFNLALADALVTTTMPFQSTVY-LMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVDVIECSLQFPDDDYDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFTDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.30 | 0.30 | 0.88 | 4.50 | Download | --------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-GAVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------- | |||||||||||||||||||
| 9 | 5o9hA | 0.25 | 0.25 | 0.85 | 3.36 | Download | ---------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYPPKVLCGVDHD-KRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARET--------------RSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR-------- | |||||||||||||||||||
| 10 | 4n6hA | 0.28 | 0.31 | 0.94 | 3.96 | Download | LEDKSPDSPTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRP-RDGAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS------KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
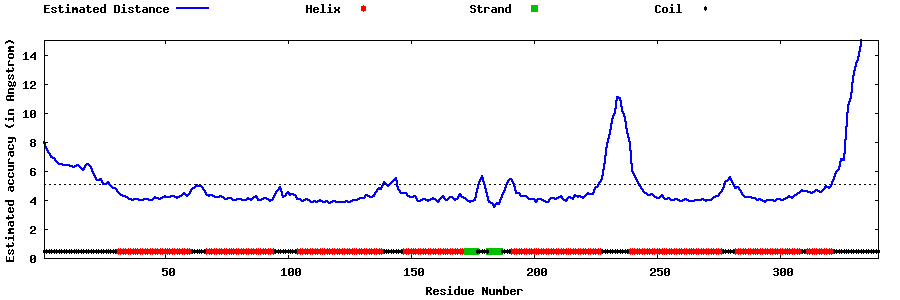
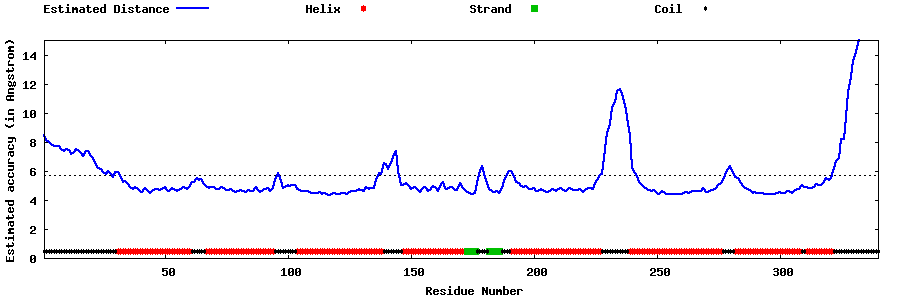
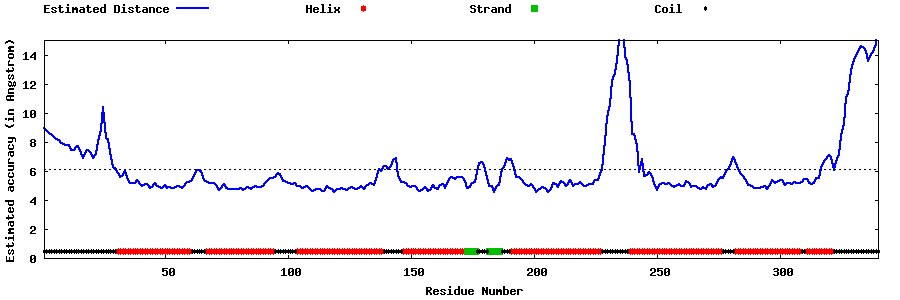
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||