

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MMKKNQTMISEFLLLGLPIQPEQQNLFYALFLAVYLTTLLGNLLVIVLIRLDSHLHMPMYLCLSNLSFSDLCFSSVTMPKLLQNMQSQNPSIPFADCLAQMYFHLFYGVLESFLLVVMAYHCYVAICFPLHYTTIMSPKCCLGLLTLSWLLTTAHATLHTLLMARLSFCAENVIPHFFCDTSTLLKLACSNTQVNGWVMFFMGGLILVIPFLLLIMSCARIVSTILRVPSTGGIQKAFSTCGPHLSVVSLFYGTIIGLYLCPLTNHNTVKDTVMAVMYTGVTHMLNPFIYSLRNRDMRGNPGQSLQHKENFFVFKIVIVGILPLLNLVGVVKLIMKYHSKSVA | |
| CCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCC | |
| 9998886175351117998821558999999999999999899999999638865562999988799875542404889988998438965738989999999999999899999999875233236754278532760899999999999999999999999741589999558936786999866164427989999999999999879999999999998840146878867610571778998999868044156078987889888189984106562434564246618899999999986089999999997368888899899999997412479 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MMKKNQTMISEFLLLGLPIQPEQQNLFYALFLAVYLTTLLGNLLVIVLIRLDSHLHMPMYLCLSNLSFSDLCFSSVTMPKLLQNMQSQNPSIPFADCLAQMYFHLFYGVLESFLLVVMAYHCYVAICFPLHYTTIMSPKCCLGLLTLSWLLTTAHATLHTLLMARLSFCAENVIPHFFCDTSTLLKLACSNTQVNGWVMFFMGGLILVIPFLLLIMSCARIVSTILRVPSTGGIQKAFSTCGPHLSVVSLFYGTIIGLYLCPLTNHNTVKDTVMAVMYTGVTHMLNPFIYSLRNRDMRGNPGQSLQHKENFFVFKIVIVGILPLLNLVGVVKLIMKYHSKSVA | |
| 8657231300200000014426212000010020122013312000000000120000000000010110110200210300100114334030200000110000002100000000011000000100200000002000000020020001001000000100010362400000012320030000102100010122133303323310220100000000102145232100000000000000111000000000021314431200000001211330010000103102300330034220000000011211230200000100131245638 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCC MMKKNQTMISEFLLLGLPIQPEQQNLFYALFLAVYLTTLLGNLLVIVLIRLDSHLHMPMYLCLSNLSFSDLCFSSVTMPKLLQNMQSQNPSIPFADCLAQMYFHLFYGVLESFLLVVMAYHCYVAICFPLHYTTIMSPKCCLGLLTLSWLLTTAHATLHTLLMARLSFCAENVIPHFFCDTSTLLKLACSNTQVNGWVMFFMGGLILVIPFLLLIMSCARIVSTILRVPSTGGIQKAFSTCGPHLSVVSLFYGTIIGLYLCPLTNHNTVKDTVMAVMYTGVTHMLNPFIYSLRNRDMRGNPGQSLQHKENFFVFKIVIVGILPLLNLVGVVKLIMKYHSKSVA | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.20 | 0.80 | 3.25 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL--------------------------------- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.21 | 0.82 | 2.05 | Download | -RGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFID-FHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-----------------GWNCEKLQSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM--------------------------------------- | |||||||||||||||||||
| 3 | 5tgzA | 0.19 | 0.21 | 0.80 | 1.80 | Download | GRGENFMD---------IENPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDRMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.23 | 0.82 | 1.51 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------ | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.21 | 0.83 | 1.24 | Download | KTTRN-AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHR-NVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------------------------------------- | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.20 | 0.81 | 3.41 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------------------------------- | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.18 | 0.77 | 1.71 | Download | ---------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----------------------------------- | |||||||||||||||||||
| 8 | 2rh1A | 0.12 | 0.22 | 0.85 | 2.56 | Download | -------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQM----HWYRATHQEAINCYAEE----TCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDEGLAKSELDKAIG--------------------------RNTNGVITKDEAEKLFNQDVDAAVRGILRNAKLKPVYDSLDAVRRAALINMVFQMGETGVAGFTNS | |||||||||||||||||||
| 9 | 3emlA | 0.21 | 0.20 | 0.81 | 4.44 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------------------------------- | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.21 | 0.88 | 5.30 | Download | -----------------S------SVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFKAAAAENLYF----------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
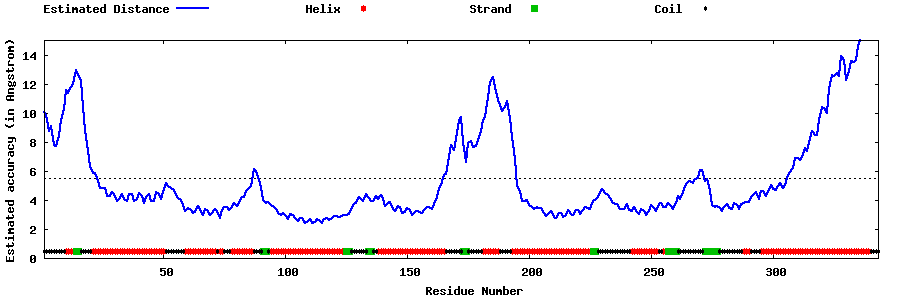
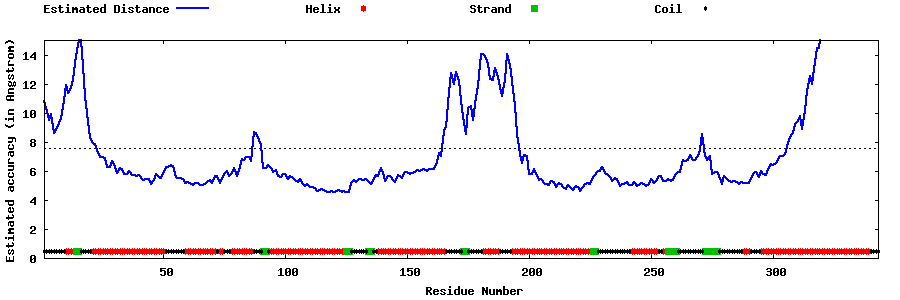
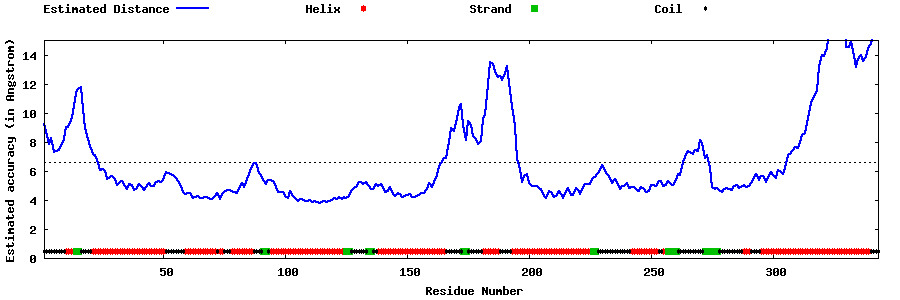
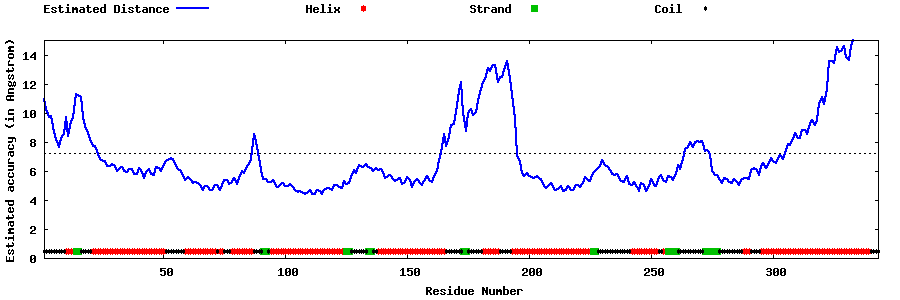
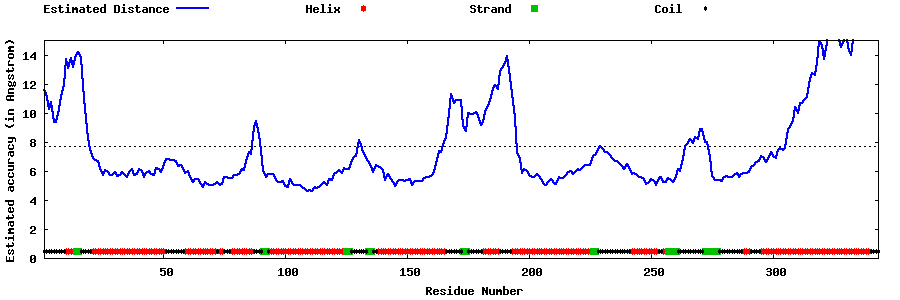
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||