

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MGDELAPCPVGTTAWPALIQLISKTPCMPQAASNTSLGLGDLRVPSSMLYWLFLPSSLLAAATLAVSPLLLVTILRNQRLRQEPHYLLPANILLSDLAYILLHMLISSSSLGGWELGRMACGILTDAVFAACTSTILSFTAIVLHTYLAVIHPLRYLSFMSHGAAWKAVALIWLVACCFPTFLIWLSKWQDAQLEEQGASYILPPSMGTQPGCGLLVIVTYTSILCVLFLCTALIANCFWRIYAEAKTSGIWGQGYSRARGTLLIHSVLITLYVSTGVVFSLDMVLTRYHHIDSGTHTWLLAANSEVLMMLPRAMLTYLYLLRYRQLLGMVRGHLPSRRHQAIFTIS | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSSCCCCCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHSSSSSSSSSSSSCCHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHCCCCHHHHHHHHCCHHHHHHHHHHCCCCCCCCSSSCC | |
| 99766789887776888656567898787765554457887666338999999999999999999999836634303656887709999999999999999989999998197657777889999999999999999999999961875521331855238999999999999999999989998525324665677732303555577775146543014356586888778775799999999999888633654306633030299999875373888888870577777674055587665664546811639999972899999999981666788865059 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MGDELAPCPVGTTAWPALIQLISKTPCMPQAASNTSLGLGDLRVPSSMLYWLFLPSSLLAAATLAVSPLLLVTILRNQRLRQEPHYLLPANILLSDLAYILLHMLISSSSLGGWELGRMACGILTDAVFAACTSTILSFTAIVLHTYLAVIHPLRYLSFMSHGAAWKAVALIWLVACCFPTFLIWLSKWQDAQLEEQGASYILPPSMGTQPGCGLLVIVTYTSILCVLFLCTALIANCFWRIYAEAKTSGIWGQGYSRARGTLLIHSVLITLYVSTGVVFSLDMVLTRYHHIDSGTHTWLLAANSEVLMMLPRAMLTYLYLLRYRQLLGMVRGHLPSRRHQAIFTIS | |
| 76442433434232113324123433333433333334264342321000010221132222033112000000011330231000000100110010101000000010223020030000000000000000001000000100000002003043312330000000000120031013010212132341443322213314233322020213101000000122021002201120010024124445434333323222002000110122223131223433425652303022023303322330002002221134012002100223334423628 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSSCCCCCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHSSSSSSSSSSSSCCHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHCCCCHHHHHHHHCCHHHHHHHHHHCCCCCCCCSSSCC MGDELAPCPVGTTAWPALIQLISKTPCMPQAASNTSLGLGDLRVPSSMLYWLFLPSSLLAAATLAVSPLLLVTILRNQRLRQEPHYLLPANILLSDLAYILLHMLISSSSLGGWELGRMACGILTDAVFAACTSTILSFTAIVLHTYLAVIHPLRYLSFMSHGAAWKAVALIWLVACCFPTFLIWLSKWQDAQLEEQGASYILPPSMGTQPGCGLLVIVTYTSILCVLFLCTALIANCFWRIYAEAKTSGIWGQGYSRARGTLLIHSVLITLYVSTGVVFSLDMVLTRYHHIDSGTHTWLLAANSEVLMMLPRAMLTYLYLLRYRQLLGMVRGHLPSRRHQAIFTIS | |||||||||||||||||||||||||
| 1 | 3emlA | 0.15 | 0.18 | 0.81 | 2.60 | Download | ---------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQN-VTNYFVVSLAAADIAVGVLAIPFAITISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLG-----WNNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQ-LVHAAKSLAIIVGLFALCWLPLHI-INCFTFFC--------PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----- | |||||||||||||||||||
| 2 | 2rh1A | 0.14 | 0.18 | 0.79 | 3.45 | Download | ----------------------------------------------VWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFIT-SLACADLVMGLAVVPFGAALMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATHQE-----AINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQ------LLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIR---KEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLC----------- | |||||||||||||||||||
| 3 | 4iaqA | 0.16 | 0.19 | 0.79 | 3.08 | Download | -------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTP-ANYLIASLAVTDLLVSILVISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPP--------FFWRQASECVVNTDHI----------LYTVYSTVGAFYFPTLLLIALYGRIYVEARSIQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHL--------AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--------- | |||||||||||||||||||
| 4 | 4ib4 | 0.15 | 0.18 | 0.81 | 1.51 | Download | ---------------------------------------EE---QGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYAT-NYFLMSLAVADLLVGLFVMPIALLTIAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKG-IET---NPNNITCVLTKE--------RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN---QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------- | |||||||||||||||||||
| 5 | 3uon | 0.16 | 0.20 | 0.80 | 1.17 | Download | ---------------------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFL-FSLACADLIIGVFSMNLYTTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP--C-IPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------- | |||||||||||||||||||
| 6 | 5tgzA | 0.15 | 0.20 | 0.83 | 2.93 | Download | ---------------------------GENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSV-------------CSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA--PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN--KLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------ | |||||||||||||||||||
| 7 | 3uon | 0.15 | 0.20 | 0.78 | 1.71 | Download | --------------------------------------------------FIVLVAGSLSLVTIIGNILVMVSIKVNRHLQ-TVNNYFLFSLACADLIIGVFSMNLYTLYIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFS------NA--AVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCA----P-CIPN-TVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------- | |||||||||||||||||||
| 8 | 2z73A | 0.14 | 0.16 | 0.79 | 1.72 | Download | ------------------------ETWWYNPSIVVHPHWREFQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQT-PANMFIINLAFSDFTFSLVPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFG------------------------WGAYTLEGVILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKR-----------------LAVVALLAQFGPLEWVT-----PYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCC--QFD | |||||||||||||||||||
| 9 | 3emlA | 0.15 | 0.18 | 0.81 | 2.62 | Download | ---------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVT-NYFVVSLAAADIAVGVLAIPFAITISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGW-----NNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAAR------RQLVHAAKSL---AIIVGLFALCWLPLHIINCFTFF-CPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----- | |||||||||||||||||||
| 10 | 3zpqA | 0.19 | 0.21 | 0.82 | 2.65 | Download | ---------------------------------------GAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLT-NLFITSLACADLVVGVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQA-----LKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQRVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP------DWLFVAFNWLGYANSAMNPIIYCR-SPDFRKAFKRLLA----------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
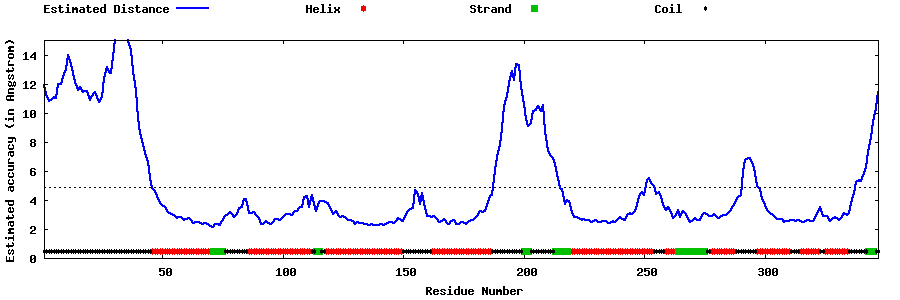 | |||
|
|
|||
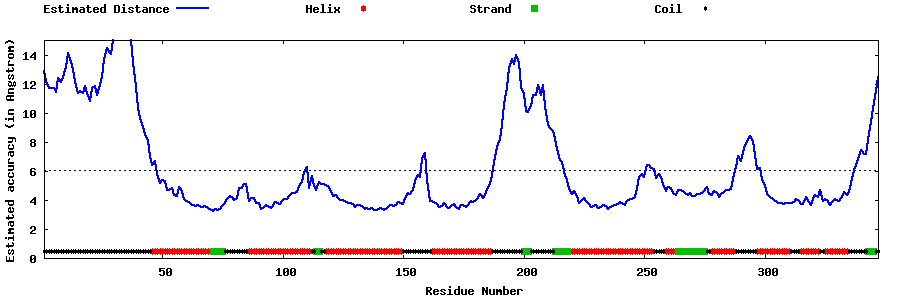 | |||
|
| |||
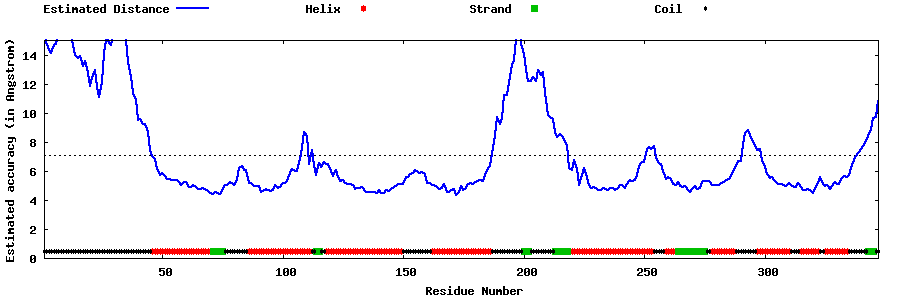 | |||
|
| |||
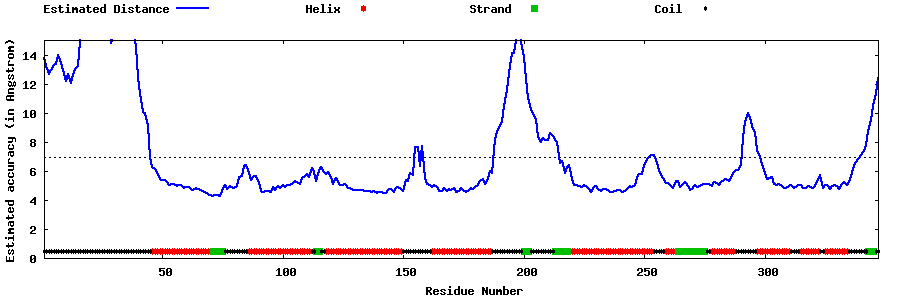 | |||
|
| |||
 | |||
|
| |||