

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MLAAAFADSNSSSMNVSFAHLHFAGGYLPSDSQDWRTIIPALLVAVCLVGFVGNLCVIGILLHNAWKGKPSMIHSLILNLSLADLSLLLFSAPIRATAYSKSVWDLGWFVCKSSDWFIHTCMAAKSLTIVVVAKVCFMYASDPAKQVSIHNYTIWSVLVAIWTVASLLPLPEWFFSTIRHHEGVEMCLVDVPAVAEEFMSMFGKLYPLLAFGLPLFFASFYFWRAYDQCKKRGTKTQNLRNQIRSKQVTVMLLSIAIISALLWLPEWVAWLWVWHLKAAGPAPPQGFIALSQVLMFSISSANPLIFLVMSEEFREGLKGVWKWMITKKPPTVSESQETPAGNSEGLPDKVPSPESPASIPEKEKPSSPSSGKGKTEKAEIPILPDVEQFWHERDTVPSVQDNDPIPWEHEDQETGEGVK | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCC | |
| 98765689888777678887667777677776009998999999999999888885410350159887886889999999999999999979999999972977895288878999999999999999999999707550042757532358889873899999999999999887899859988997408962879999999999999999999999999999999982367875145667751357688899999999999999999999996020241189999999999999999999999997499999999999876157888876666678788778888767899999888767789997778887655556779985312213789884001788960203665577889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MLAAAFADSNSSSMNVSFAHLHFAGGYLPSDSQDWRTIIPALLVAVCLVGFVGNLCVIGILLHNAWKGKPSMIHSLILNLSLADLSLLLFSAPIRATAYSKSVWDLGWFVCKSSDWFIHTCMAAKSLTIVVVAKVCFMYASDPAKQVSIHNYTIWSVLVAIWTVASLLPLPEWFFSTIRHHEGVEMCLVDVPAVAEEFMSMFGKLYPLLAFGLPLFFASFYFWRAYDQCKKRGTKTQNLRNQIRSKQVTVMLLSIAIISALLWLPEWVAWLWVWHLKAAGPAPPQGFIALSQVLMFSISSANPLIFLVMSEEFREGLKGVWKWMITKKPPTVSESQETPAGNSEGLPDKVPSPESPASIPEKEKPSSPSSGKGKTEKAEIPILPDVEQFWHERDTVPSVQDNDPIPWEHEDQETGEGVK | |
| 63434133432222020223243343222232210000002111200110130110000000113244110000000200010000000001000000001321100200000000000000000000000000000000000022241121000000000010000000000000202437312000000134333010000001011011200100020001000102334444544443343100000000000000000212000000000142233201000100000000100000000000004300410220021023343344443453444445435443434444443455444443454644446343331353542344364353466431232435546456368 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCSSSSSSCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCC MLAAAFADSNSSSMNVSFAHLHFAGGYLPSDSQDWRTIIPALLVAVCLVGFVGNLCVIGILLHNAWKGKPSMIHSLILNLSLADLSLLLFSAPIRATAYSKSVWDLGWFVCKSSDWFIHTCMAAKSLTIVVVAKVCFMYASDPAKQVSIHNYTIWSVLVAIWTVASLLPLPEWFFSTIRHHEGVEMCLVDVPAVAEEFMSMFGKLYPLLAFGLPLFFASFYFWRAYDQCKKRGTKTQNLRNQIRSKQVTVMLLSIAIISALLWLPEWVAWLWVWHLKAAGPAPPQGFIALSQVLMFSISSANPLIFLVMSEEFREGLKGVWKWMITKKPPTVSESQETPAGNSEGLPDKVPSPESPASIPEKEKPSSPSSGKGKTEKAEIPILPDVEQFWHERDTVPSVQDNDPIPWEHEDQETGEGVK | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.18 | 0.16 | 0.71 | 2.92 | Download | -----------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVR--YTKMKTATNIYIFNLALADALATSTLPFQSAKYLM--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 4n6hA | 0.19 | 0.17 | 0.68 | 3.64 | Download | ----------------------------------LAIAITALYSAVCAVGLLGNVLVMFGIVR--YTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 3 | 4n6hA | 0.18 | 0.16 | 0.72 | 3.55 | Download | -----------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.14 | 0.20 | 0.84 | 1.51 | Download | ----DLR-DNETWWYNPSIIVHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQ--TPANMFIINLAFSDFTFSLVNFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGYTLEGVLCNCSFDYISRDS-TTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLE--WVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDA-ETEIPAGESSDA--------------APSADAAQMKE---------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.18 | 0.18 | 0.68 | 1.16 | Download | -------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 6 | 4n6hA | 0.18 | 0.16 | 0.71 | 3.32 | Download | -------------------------GARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.17 | 0.18 | 0.67 | 1.70 | Download | ---------------------------------AIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS-----AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.18 | 0.16 | 0.72 | 4.42 | Download | ----------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.18 | 0.16 | 0.72 | 3.14 | Download | -----------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVR--YTKMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.18 | 0.17 | 0.70 | 3.58 | Download | --------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVR--YTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------------------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
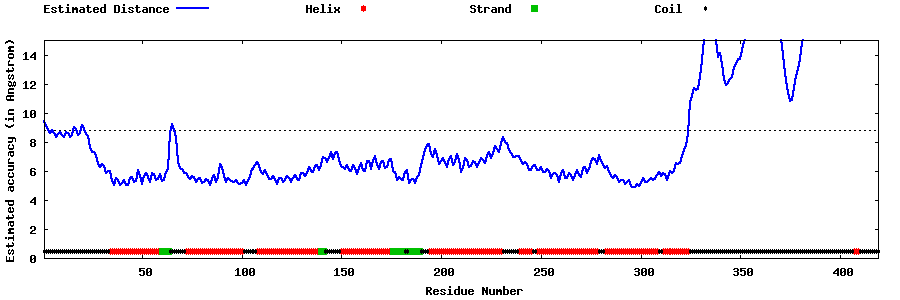
| Generated 3D models | Estimated local accuracy of models | ||
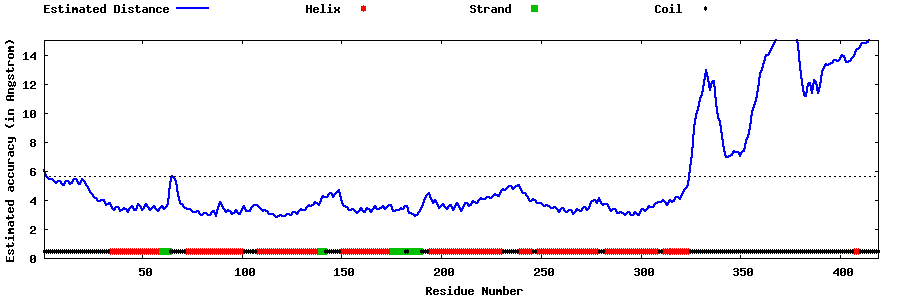 | |||
|
|
|||
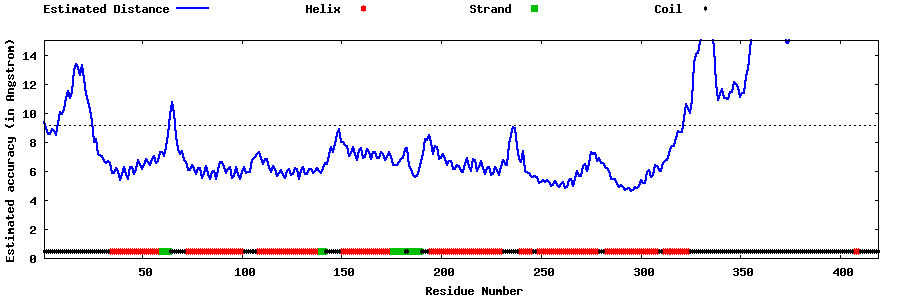 | |||
|
| |||
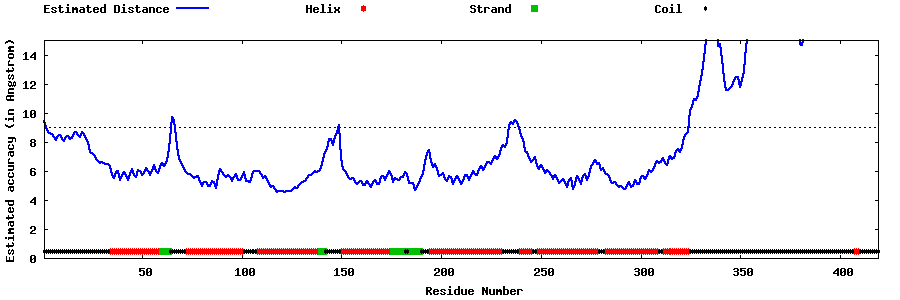 | |||
|
| |||
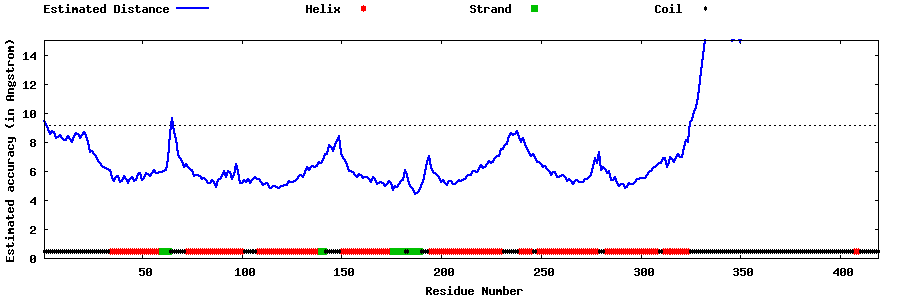 | |||
|
| |||
 | |||
|
| |||