

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MPTLNTSASPPTFFWANASGGSVLSADDAPMPVKFLALRLMVALAYGLVGAIGLLGNLAVLWVLSNCARRAPGPPSDTFVFNLALADLGLALTLPFWAAESALDFHWPFGGALCKMVLTATVLNVYASIFLITALSVARYWVVAMAAGPGTHLSLFWARIATLAVWAAAALVTVPTAVFGVEGEVCGVRLCLLRFPSRYWLGAYQLQRVVLAFMVPLGVITTSYLLLLAFLQRRQRRRQDSRVVARSVRILVASFFLCWFPNHVVTLWGVLVKFDLVPWNSTFYTIQTYVFPVTTCLAHSNSCLNPVLYCLLRREPRQALAGTFRDLRLRLWPQGGGWVQQVALKQVGRRWVASNPRESRPSTLLTNLDRGTPG | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99987899987667678888877787766664220158998999999999999987997400100225667886999999999999999999849999999769987678778999999999999999999999999836320112763313360446330399999999999799886146813990687087991789999999999999999999999999999999851515450233567888899999999857999999999999837788876799999999999999999999798999996589899999999886500533677887667502567888788898757887766778899988 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MPTLNTSASPPTFFWANASGGSVLSADDAPMPVKFLALRLMVALAYGLVGAIGLLGNLAVLWVLSNCARRAPGPPSDTFVFNLALADLGLALTLPFWAAESALDFHWPFGGALCKMVLTATVLNVYASIFLITALSVARYWVVAMAAGPGTHLSLFWARIATLAVWAAAALVTVPTAVFGVEGEVCGVRLCLLRFPSRYWLGAYQLQRVVLAFMVPLGVITTSYLLLLAFLQRRQRRRQDSRVVARSVRILVASFFLCWFPNHVVTLWGVLVKFDLVPWNSTFYTIQTYVFPVTTCLAHSNSCLNPVLYCLLRREPRQALAGTFRDLRLRLWPQGGGWVQQVALKQVGRRWVASNPRESRPSTLLTNLDRGTPG | |
| 65443343334423231323333233442434332200000002201200220232121001000103323431000000000010201000000000000025342100100010010112211100000000001000000000020232112100100000011000000100000020353642210102023600210010111230333122000200010011023146444443000000000000010031100000001002304103342322200100100000001200112000100004401410140034003322564443245443554444444344454443341342565458 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MPTLNTSASPPTFFWANASGGSVLSADDAPMPVKFLALRLMVALAYGLVGAIGLLGNLAVLWVLSNCARRAPGPPSDTFVFNLALADLGLALTLPFWAAESALDFHWPFGGALCKMVLTATVLNVYASIFLITALSVARYWVVAMAAGPGTHLSLFWARIATLAVWAAAALVTVPTAVFGVEGEVCGVRLCLLRFPSRYWLGAYQLQRVVLAFMVPLGVITTSYLLLLAFLQRRQRRRQDSRVVARSVRILVASFFLCWFPNHVVTLWGVLVKFDLVPWNSTFYTIQTYVFPVTTCLAHSNSCLNPVLYCLLRREPRQALAGTFRDLRLRLWPQGGGWVQQVALKQVGRRWVASNPRESRPSTLLTNLDRGTPG | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.25 | 0.22 | 0.78 | 3.22 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINY--KRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.25 | 0.23 | 0.78 | 3.88 | Download | ----------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILIN--YKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLREVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.31 | 0.26 | 0.79 | 3.75 | Download | --------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMK--TATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVD------IDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG--------------------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.19 | 0.23 | 0.93 | 1.51 | Download | --DLRDNETW----WYNPSIIV--HPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT--KSLQTPANMFIINLAFSDFTFLVGFPLMTISCF-LKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYLEGVLCNCSFDYISRSTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV---------TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDKETEDDKDAETEIPA--GESSDAAPSADAA-QMKE---- | |||||||||||||||||||
| 5 | 5t1a | 0.24 | 0.25 | 0.76 | 1.16 | Download | ---------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILIN--CKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQK-EDSVYVCGPYFPRG-WNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFG-LSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF-------------------------------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.25 | 0.22 | 0.78 | 3.32 | Download | ----------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYK--RLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------- | |||||||||||||||||||
| 7 | 2ziy | 0.22 | 0.23 | 0.76 | 1.67 | Download | -------------------------------DQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT--KSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFWGAYTLEGVLCNCSFDYISRDTTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEW---------VTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVL---------------------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.31 | 0.26 | 0.79 | 4.44 | Download | -------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY--TKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQFPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV--DIDRRD----PLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK-PCG-------------------------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.25 | 0.22 | 0.78 | 3.48 | Download | ---------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINY--KRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYQFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLN-NCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.28 | 0.23 | 0.77 | 4.08 | Download | -----------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYT--KMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET-------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
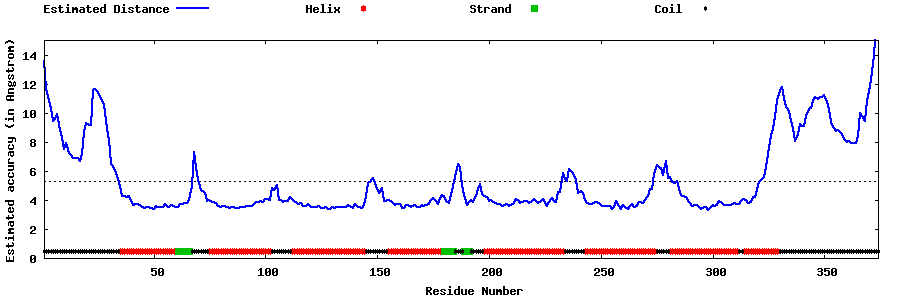
| Generated 3D models | Estimated local accuracy of models | ||
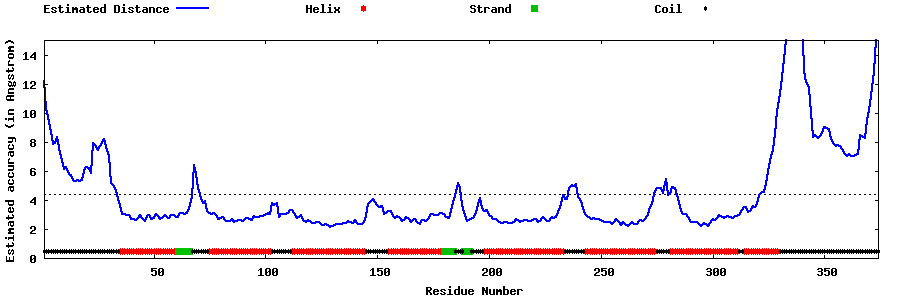 | |||
|
|
|||
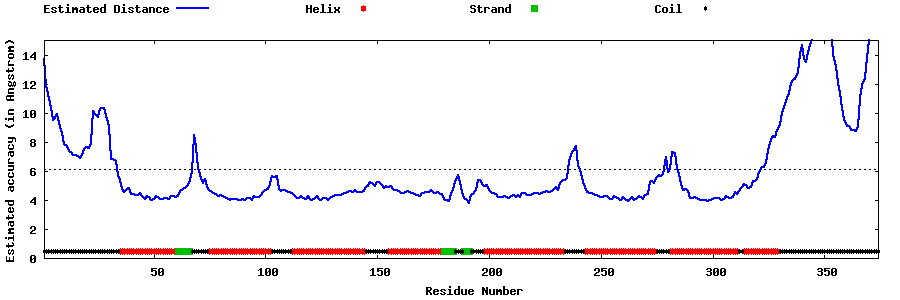 | |||
|
| |||
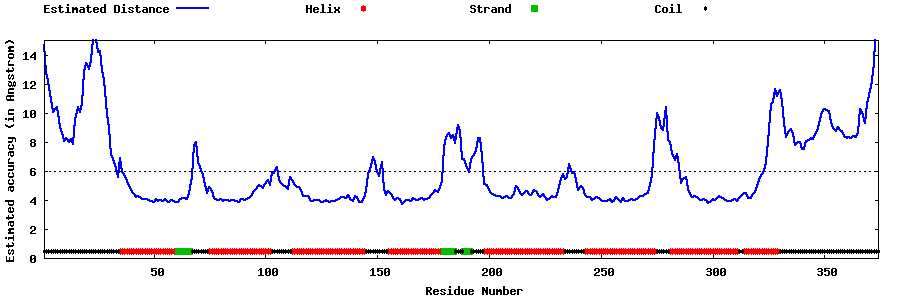 | |||
|
| |||
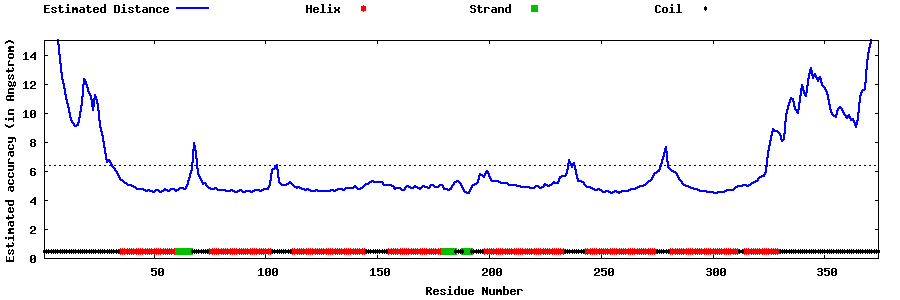 | |||
|
| |||
 | |||
|
| |||