

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNQTLNSSGTVESALNYSRGSTVHTAYLVLSSLAMFTCLCGMAGNSMVIWLLGFRMHRNPFCIYILNLAAADLLFLFSMASTLSLETQPLVNTTDKVHELMKRLMYFAYTVGLSLLTAISTQRCLSVLFPIWFKCHRPRHLSAWVCGLLWTLCLLMNGLTSSFCSKFLKFNEDRCFRVDMVQAALIMGVLTPVMTLSSLTLFVWVRRSSQQWRRQPTRLFVVVLASVLVFLICSLPLSIYWFVLYWLSLPPEMQVLCFSLSRLSSSVSSSANPVIYFLVGSRRSHRLPTRSLGTVLQQALREEPELEGGETPTVGTNEMGA | |
| CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHCHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCC | |
| 999888899988888888777511999999999999999999989998998873478977599999999999999999999999997386784349999999999999999999999999999999999987146787147999999999999999858999811104799860668999999999988999999999999999997155468778508999999999999987899999999999845724999999999999999998876976256888868986431899975252267888999999998787889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MNQTLNSSGTVESALNYSRGSTVHTAYLVLSSLAMFTCLCGMAGNSMVIWLLGFRMHRNPFCIYILNLAAADLLFLFSMASTLSLETQPLVNTTDKVHELMKRLMYFAYTVGLSLLTAISTQRCLSVLFPIWFKCHRPRHLSAWVCGLLWTLCLLMNGLTSSFCSKFLKFNEDRCFRVDMVQAALIMGVLTPVMTLSSLTLFVWVRRSSQQWRRQPTRLFVVVLASVLVFLICSLPLSIYWFVLYWLSLPPEMQVLCFSLSRLSSSVSSSANPVIYFLVGSRRSHRLPTRSLGTVLQQALREEPELEGGETPTVGTNEMGA | |
| 714233443334433433434323011021132132213333333010000010234310100000000103001100100100110233111000000001221331121102000000200010000002024213311000000101210010010000000012454422001000112122312120112002011001010224566444300000000001001002232211000100132333001001100101012301220000000034014303430032004300434444676644434465468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHCHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCC MNQTLNSSGTVESALNYSRGSTVHTAYLVLSSLAMFTCLCGMAGNSMVIWLLGFRMHRNPFCIYILNLAAADLLFLFSMASTLSLETQPLVNTTDKVHELMKRLMYFAYTVGLSLLTAISTQRCLSVLFPIWFKCHRPRHLSAWVCGLLWTLCLLMNGLTSSFCSKFLKFNEDRCFRVDMVQAALIMGVLTPVMTLSSLTLFVWVRRSSQQWRRQPTRLFVVVLASVLVFLICSLPLSIYWFVLYWLSLPPEMQVLCFSLSRLSSSVSSSANPVIYFLVGSRRSHRLPTRSLGTVLQQALREEPELEGGETPTVGTNEMGA | |||||||||||||||||||||||||
| 1 | 5o9hA | 0.22 | 0.25 | 0.88 | 3.24 | Download | --------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPVLCGVRAVAIVRLVLGFLWPLLTLTICYTFILLRTW-SARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------- | |||||||||||||||||||
| 2 | 4ntjA | 0.14 | 0.16 | 0.79 | 3.91 | Download | -------------------TRDYKITQVLFPLLYTVLFFVGLITNGLAMRIFFQIRSKSNFIIFLKNTVISDLLMILTFPFKILSD---AKLLRTFVCQVTSVIFYFTMYISISFLGLITIDRYQKTTRPF---KTKNLLGAKILSVVIWAFMFLLSLPNMIL------KSEFGLVWHEIVNYICQVIFWINFLIVIVCYTLITKEL-----YRSRKKVNVKVFIIIAVFFICFVPFHFARIPYTLSQTRENTLFYVKESTLWLTSLNACLNPFIYFFLCKSFRNSLISM------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.17 | 0.22 | 0.88 | 3.58 | Download | --------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYKMKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.16 | 0.22 | 0.85 | 1.51 | Download | --------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLKKLQYATNYFLMSLAVADLLVGLFVMPIALLTMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIET-NPNNITCVLFLFGSLAAF-FTPLAIMIVTYFLTIHALQKKAADTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAF-----GRYITCNYR-------------------- | |||||||||||||||||||
| 5 | 4djh | 0.19 | 0.16 | 0.83 | 1.15 | Download | ----------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA----LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------------------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.23 | 0.25 | 0.87 | 3.22 | Download | ---------------------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYDHDKRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSAR-ETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------- | |||||||||||||||||||
| 7 | 3uon | 0.16 | 0.21 | 0.81 | 1.69 | Download | --------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKNRHLQTVNNYFLFSLACADLIIGVFMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVTVEDGECYNAAVTFGTAIAAFYLPVIIMTVLYWHISRNIDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP--CIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKH-------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.16 | 0.22 | 0.88 | 3.80 | Download | -------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKKTATNIYIFNLALADALATSTLPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQ-----LCRKPCG-------------------- | |||||||||||||||||||
| 9 | 5o9hA | 0.22 | 0.25 | 0.88 | 3.26 | Download | --------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEAKRTINAIWFLNLAVADFLACLALPALFTSIVQHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSAR-ETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLEPSSLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR------------------- | |||||||||||||||||||
| 10 | 6b73A | 0.20 | 0.22 | 0.83 | 4.31 | Download | ------------------LGSISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLCSLQFPDDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRSRRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGTSHSTAALSSYYFCIALGYTNSSLNPILYAFLDENFKR------------------------------------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
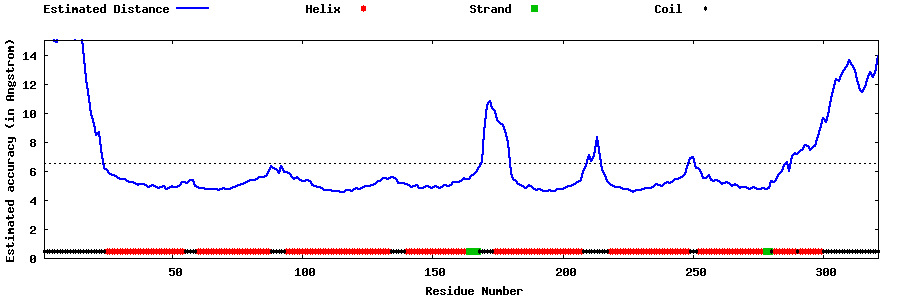
| Generated 3D models | Estimated local accuracy of models | ||
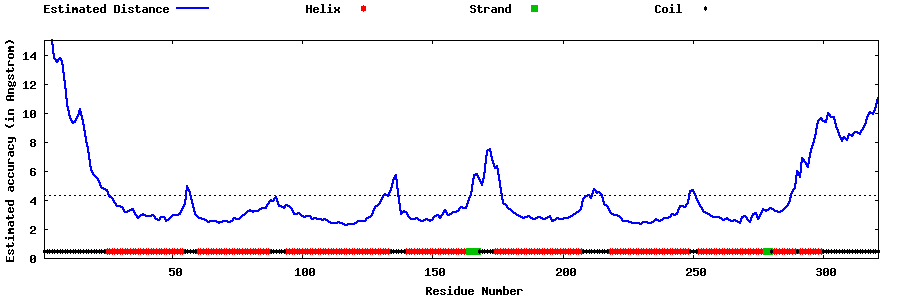 | |||
|
|
|||
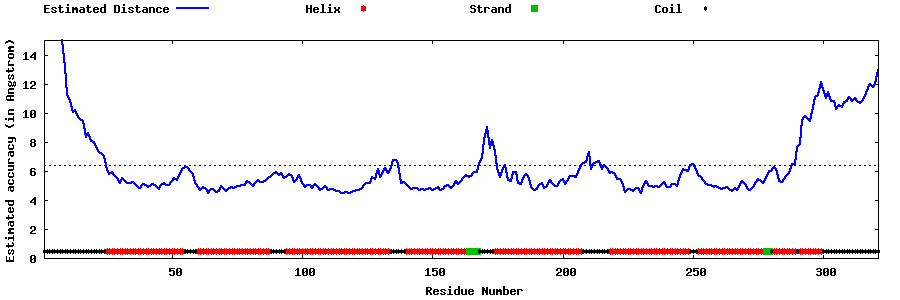 | |||
|
| |||
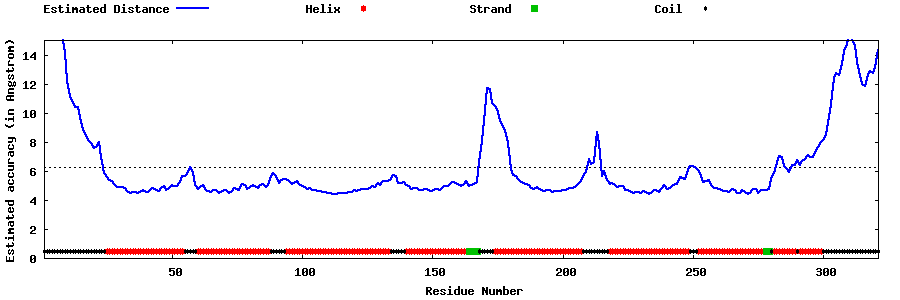 | |||
|
| |||
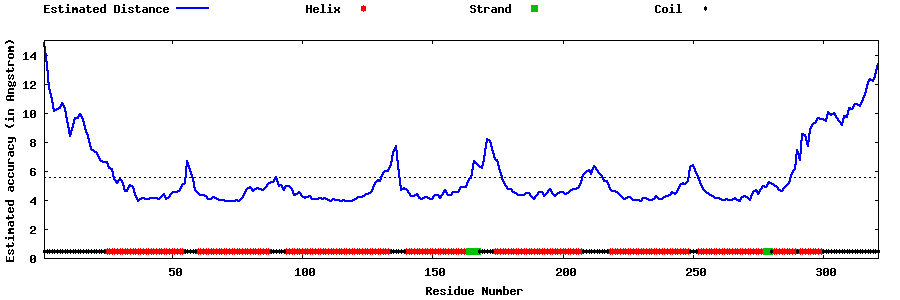 | |||
|
| |||
 | |||
|
| |||