

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MLCHRGGQLIVPIIPLCPEHSCRGRRLQNLLSGPWPKQPMELHNLSSPSPSLSSSVLPPSFSPSPSSAPSAFTTVGGSSGGPCHPTSSSLVSAFLAPILALEFVLGLVGNSLALFIFCIHTRPWTSNTVFLVSLVAADFLLISNLPLRVDYYLLHETWRFGAAACKVNLFMLSTNRTASVVFLTAIALNRYLKVVQPHHVLSRASVGAAARVAGGLWVGILLLNGHLLLSTFSGPSCLSYRVGTKPSASLRWHQALYLLEFFLPLALILFAIVSIGLTIRNRGLGGQAGPQRAMRVLAMVVAVYTICFLPSIIFGMASMVAFWLSACRSLDLCTQLFHGSLAFTYLNSVLDPVLYCFSSPNFLHQSRALLGLTRGRQGPVSDESSYQPSRQWRYREASRKAEAIGKLKVQGEVSLEKEGSSQG | |
| CCCCCCCCSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 954567644113366786666666510014579888887531125899998765558888999988888777777887788776530778999999999999999998899855743226777858899999999999999999899999996589987876889999999999999999999999997235301215245453645576656899999999987885485346643664278884089999999999989999999999999999998387677764335389999999999999847999999999998633664089999999999999999999788999993188999999999562013678877666767787877357777644225666788875566788898 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MLCHRGGQLIVPIIPLCPEHSCRGRRLQNLLSGPWPKQPMELHNLSSPSPSLSSSVLPPSFSPSPSSAPSAFTTVGGSSGGPCHPTSSSLVSAFLAPILALEFVLGLVGNSLALFIFCIHTRPWTSNTVFLVSLVAADFLLISNLPLRVDYYLLHETWRFGAAACKVNLFMLSTNRTASVVFLTAIALNRYLKVVQPHHVLSRASVGAAARVAGGLWVGILLLNGHLLLSTFSGPSCLSYRVGTKPSASLRWHQALYLLEFFLPLALILFAIVSIGLTIRNRGLGGQAGPQRAMRVLAMVVAVYTICFLPSIIFGMASMVAFWLSACRSLDLCTQLFHGSLAFTYLNSVLDPVLYCFSSPNFLHQSRALLGLTRGRQGPVSDESSYQPSRQWRYREASRKAEAIGKLKVQGEVSLEKEGSSQG | |
| 622333231111010204424144451443343623433143541633423353332333232333333332232334333313333330110000210120023123232200100002234220000000000000000000000000000244301001000000000112111000000000010000000000202321322000000000010000000000000333330211022433312000100101211331120002000000000233535644421000000000000000012200100001001311530422300100110010000200020000000006600510140032234364544463344444444444444444335544343434345664678 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MLCHRGGQLIVPIIPLCPEHSCRGRRLQNLLSGPWPKQPMELHNLSSPSPSLSSSVLPPSFSPSPSSAPSAFTTVGGSSGGPCHPTSSSLVSAFLAPILALEFVLGLVGNSLALFIFCIHTRPWTSNTVFLVSLVAADFLLISNLPLRVDYYLLHETWRFGAAACKVNLFMLSTNRTASVVFLTAIALNRYLKVVQPHHVLSRASVGAAARVAGGLWVGILLLNGHLLLSTFSGPSCLSYRVGTKPSASLRWHQALYLLEFFLPLALILFAIVSIGLTIRNRGLGGQAGPQRAMRVLAMVVAVYTICFLPSIIFGMASMVAFWLSACRSLDLCTQLFHGSLAFTYLNSVLDPVLYCFSSPNFLHQSRALLGLTRGRQGPVSDESSYQPSRQWRYREASRKAEAIGKLKVQGEVSLEKEGSSQG | |||||||||||||||||||||||||
| 1 | 5unfA | 0.18 | 0.21 | 0.86 | 2.86 | Download | NDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAHLDAIPILYYIIFVIGFLVNIVVVTLFCCQKGPKKVSSIYIFNLAVADLLLLATLPLWATYYSYRYDWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVIYPFP-------WQASYIVPLVWCMACLSSLPTFYFRDVRNACIMAFPPEKAQWSAGIALMKNILGFIIPLIFIATCYFGIRKHLLKTNSYGKITRDQVLKMAAAVVLAFIICWLPFHVLTFLDALAWMINSCEVIAVIDLALPFAILLGFTNSCVNPFLYCFVGNRFQQKLRSVFR---------------------------------------------------- | |||||||||||||||||||
| 2 | 4yayA | 0.22 | 0.19 | 0.68 | 3.82 | Download | --------------------------------------------------------------------------LILNSSDCPKAGRHNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFFI-ITVCAFHYETLPIGLGLTKNILGFLFPFLIILTSYTLIWKALKK--------NDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQLIRDCRIADIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.20 | 0.22 | 0.87 | 3.47 | Download | -LTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRR---DPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 4 | 4n6hA | 0.25 | 0.20 | 0.69 | 3.52 | Download | -------------------------------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV---DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 5 | 4yayA | 0.18 | 0.19 | 0.85 | 3.25 | Download | KMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLILNSSDCPKAGRHNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFFI-ITVCAFHYETLPIGLGLTKNILGFLFPFLIILTSYTLIWKALKK--------NDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQLIRDCRIADIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----------------------------------------------------- | |||||||||||||||||||
| 6 | 4djh | 0.24 | 0.20 | 0.65 | 1.52 | Download | ---------------------------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMN-SWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTVIECSLQFPDDDYSWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLEKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA---------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------- | |||||||||||||||||||
| 7 | 4n6hA | 0.22 | 0.22 | 0.88 | 3.04 | Download | VIEKADNDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSRLLSGSKEKRRITRMVLVVVGAFVVCWAPIHIFVIVWTL---VDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------ | |||||||||||||||||||
| 8 | 5t1a | 0.20 | 0.17 | 0.67 | 1.17 | Download | --------------------------------------------------------------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKEDSVYVCGPYFPGWNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF----------------------------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.21 | 0.22 | 0.88 | 4.14 | Download | DLIEKADNALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLME-TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGSKLRRITRMVLVVVGAFVVCWAPIHIFVIVWTL---VDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK------------------------------------------------PCG | |||||||||||||||||||
| 10 | 4djh | 0.24 | 0.20 | 0.64 | 1.70 | Download | ------------------------------------------------------------------------------------------IPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVY-LMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKDVDVIECSLQFPDDDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---A------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF---------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
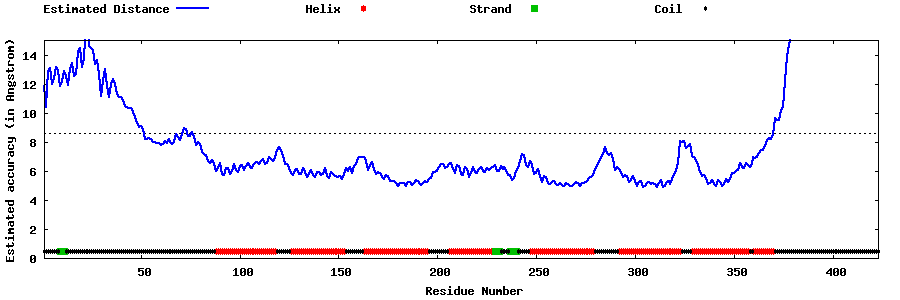
| Generated 3D models | Estimated local accuracy of models | ||
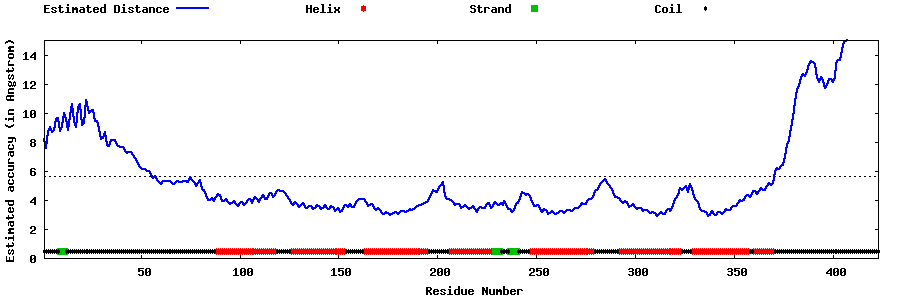 | |||
|
|
|||
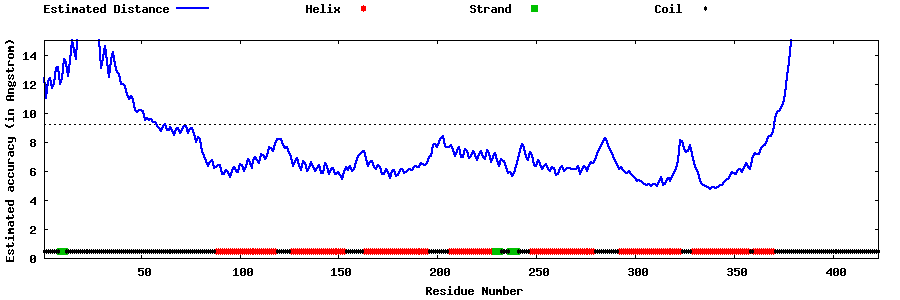 | |||
|
| |||
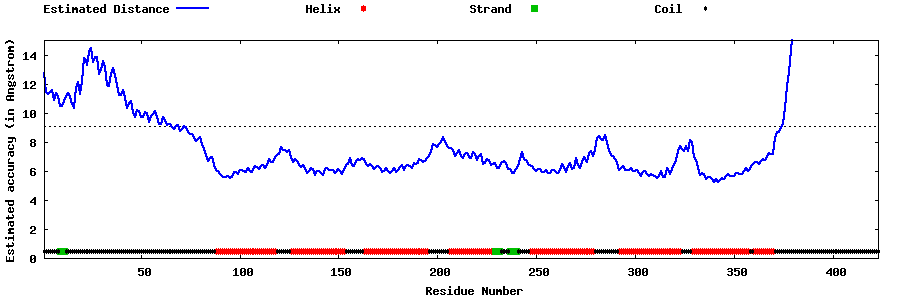 | |||
|
| |||
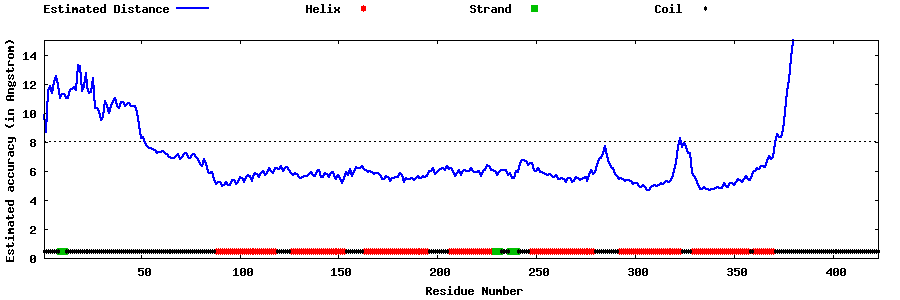 | |||
|
| |||
 | |||
|
| |||