

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| METTMGFMDDNATNTSTSFLSVLNPHGAHATSFPFNFSYSDYDMPLDEDEDVTNSRTFFAAKIVIGMALVGIMLVCGIGNFIFIAALVRYKKLRNLTNLLIANLAISDFLVAIVCCPFEMDYYVVRQLSWEHGHVLCTSVNYLRTVSLYVSTNALLAIAIDRYLAIVHPLRPRMKCQTATGLIALVWTVSILIAIPSAYFTTETVLVIVKSQEKIFCGQIWPVDQQLYYKSYFLFIFGIEFVGPVVTMTLCYARISRELWFKAVPGFQTEQIRKRLRCRRKTVLVLMCILTAYVLCWAPFYGFTIVRDFFPTVFVKEKHYLTAFYIVECIAMSNSMINTLCFVTVKNDTVKYFKKIMLLHWKASYNGGKSSADLDLKTIGMPATEEVDCIRLK | |
| CCCCHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 997400003688888765456689888878788887787655678776544444554107999999999999999989999885887528988854238999999999999999828999999983898226558988899999999999999999999996999764177540041587732699999999999999992415743207898069983389724799999999999999999999999999999999816689986526678988864579999999999999997799999999996522245506999999999999999999999999980999999999985143268888888877778774788999997765579 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| METTMGFMDDNATNTSTSFLSVLNPHGAHATSFPFNFSYSDYDMPLDEDEDVTNSRTFFAAKIVIGMALVGIMLVCGIGNFIFIAALVRYKKLRNLTNLLIANLAISDFLVAIVCCPFEMDYYVVRQLSWEHGHVLCTSVNYLRTVSLYVSTNALLAIAIDRYLAIVHPLRPRMKCQTATGLIALVWTVSILIAIPSAYFTTETVLVIVKSQEKIFCGQIWPVDQQLYYKSYFLFIFGIEFVGPVVTMTLCYARISRELWFKAVPGFQTEQIRKRLRCRRKTVLVLMCILTAYVLCWAPFYGFTIVRDFFPTVFVKEKHYLTAFYIVECIAMSNSMINTLCFVTVKNDTVKYFKKIMLLHWKASYNGGKSSADLDLKTIGMPATEEVDCIRLK | |
| 743513313332233333222313333323132321202322423244344233231120000000100120020023132000000001330210011002001201000000001010000003333010010001001000000000000000000000010001002332333100000000010001001000000202314234443110010201344420100000010221113101000200010001023342324445434433443200000000000000000213000000100134213422000001000000000000000000000043004002300100223444555444434343343444543434338 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSSCCCCCSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC METTMGFMDDNATNTSTSFLSVLNPHGAHATSFPFNFSYSDYDMPLDEDEDVTNSRTFFAAKIVIGMALVGIMLVCGIGNFIFIAALVRYKKLRNLTNLLIANLAISDFLVAIVCCPFEMDYYVVRQLSWEHGHVLCTSVNYLRTVSLYVSTNALLAIAIDRYLAIVHPLRPRMKCQTATGLIALVWTVSILIAIPSAYFTTETVLVIVKSQEKIFCGQIWPVDQQLYYKSYFLFIFGIEFVGPVVTMTLCYARISRELWFKAVPGFQTEQIRKRLRCRRKTVLVLMCILTAYVLCWAPFYGFTIVRDFFPTVFVKEKHYLTAFYIVECIAMSNSMINTLCFVTVKNDTVKYFKKIMLLHWKASYNGGKSSADLDLKTIGMPATEEVDCIRLK | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.21 | 0.20 | 0.76 | 2.85 | Download | -------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM----TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVD-IDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.22 | 0.21 | 0.75 | 3.82 | Download | ------------------------------------------------------------FKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAED--WPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRGIGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMD-YKGSYLRICLLHPVQKTAFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNSFLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC---------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.20 | 0.76 | 3.53 | Download | -------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMET--WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMA-----VTRPRDGAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLV-DIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 4 | 2ziy | 0.19 | 0.23 | 0.89 | 1.53 | Download | ---------DLRDNE--------------------TWWYNPSII--VHPHWREFDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVGFPLMTISCFL--KKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTL----EGVLCNCSFDYISRD-STTRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV----TPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCFDDKETDAETEIPAGESSDAAPSADAAQMK | |||||||||||||||||||
| 5 | 4rnb | 0.27 | 0.23 | 0.74 | 1.15 | Download | ---------------------------------------------------------PKEYEWVLIAGYIIVFVVALIGNVLVCVAVWKNHHMRTVTNYFIVNLSLADVLVTITCLPATLVVDITE--TWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPST----AKRARNSIVIIWIVSCIIMIPQAIVMECSTVF--KTTLFTVCDERWGGE--IYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSLSSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMFAHDRETVYAWFTFSHWLVYANSAANPIIYNFLSGKFREEFKAAFSC---------------------------------- | |||||||||||||||||||
| 6 | 4n6hA | 0.20 | 0.21 | 0.87 | 3.12 | Download | -DILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLGSPG------ARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALA-TSTLPFQSAKYLMET--WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLS----GSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI-DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.23 | 0.20 | 0.72 | 1.70 | Download | -------------------------------------------------------------VVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTV--IGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGV--RTVEDGECYIQFFSN-----AAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFGAAWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCI-----PNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------------------------------- | |||||||||||||||||||
| 8 | 4n6hA | 0.21 | 0.20 | 0.77 | 4.64 | Download | ------------------------------------------------GSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLME--TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDI-DRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 9 | 4n6hA | 0.21 | 0.20 | 0.76 | 2.95 | Download | -------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATSTLPFQSAKYLM----TWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRD-----GAVVCMLQFPSPSWYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSGS----KEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVD-IDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.21 | 0.20 | 0.75 | 3.28 | Download | ----------------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLM--GTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYR-----QGSIDCTLTFSHPTWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGS----KEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALIT--IPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF---------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
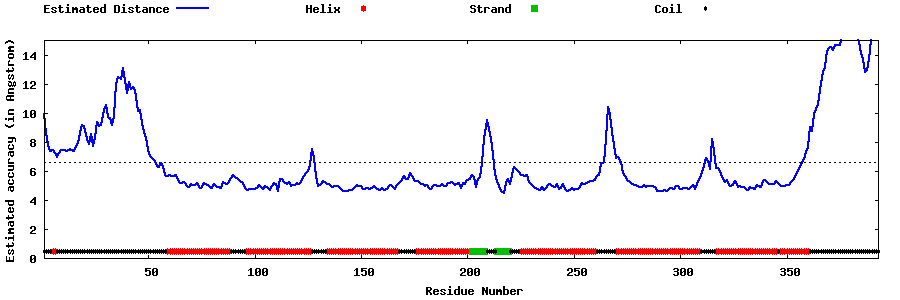
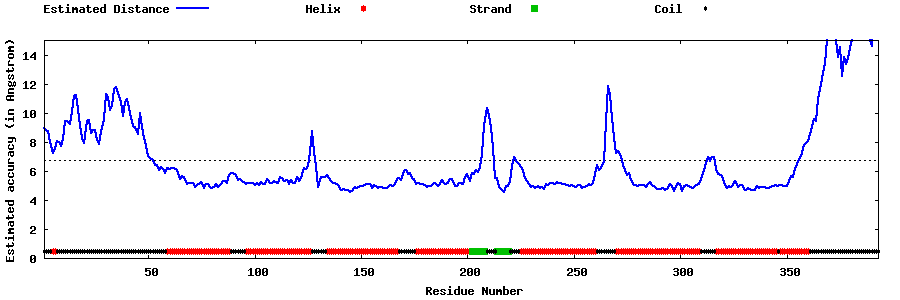
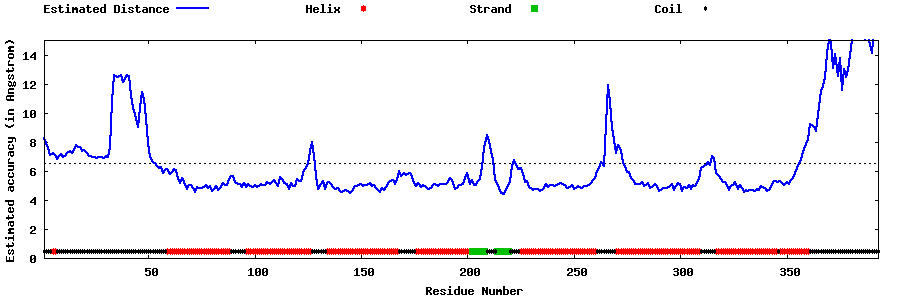
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||