

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MVDPNGNESSATYFILIGLPGLEEAQFWLAFPLCSLYLIAVLGNLTIIYIVRTEHSLHEPMYIFLCMLSGIDILISTSSMPKMLAIFWFNSTTIQFDACLLQMFAIHSLSGMESTVLLAMAFDRYVAICHPLRHATVLTLPRVTKIGVAAVVRGAALMAPLPVFIKQLPFCRSNILSHSYCLHQDVMKLACDDIRVNVVYGLIVIISAIGLDSLLISFSYLLILKTVLGLTREAQAKAFGTCVSHVCAVFIFYVPFIGLSMVHRFSKRRDSPLPVILANIYLLVPPVLNPIVYGVKTKEIRQRILRLFHVATHASEP | |
| CCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCCCCCC | |
| 99989997753323057998986789999999999999999999999999856887542089999999999999999659999999956998688888999999999999999999999983427862666456412888999999999999999999989999961899998936861111276788834783587899999999999998999999999999999804998888987616999999999999999997202357989947999999999627766468011065799999999983277767997 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MVDPNGNESSATYFILIGLPGLEEAQFWLAFPLCSLYLIAVLGNLTIIYIVRTEHSLHEPMYIFLCMLSGIDILISTSSMPKMLAIFWFNSTTIQFDACLLQMFAIHSLSGMESTVLLAMAFDRYVAICHPLRHATVLTLPRVTKIGVAAVVRGAALMAPLPVFIKQLPFCRSNILSHSYCLHQDVMKLACDDIRVNVVYGLIVIISAIGLDSLLISFSYLLILKTVLGLTREAQAKAFGTCVSHVCAVFIFYVPFIGLSMVHRFSKRRDSPLPVILANIYLLVPPVLNPIVYGVKTKEIRQRILRLFHVATHASEP | |
| 64442434231310200000121320200011022012203321210000020141001000200000020010003002020000000214503140000001200310331110011002000000021010000003410010000002212011202101021031045200000001021002000230300111001003313333231032011100200020046013301200100000000032213310100100330011000110221003003300300002034004100300233463668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCCCCCC MVDPNGNESSATYFILIGLPGLEEAQFWLAFPLCSLYLIAVLGNLTIIYIVRTEHSLHEPMYIFLCMLSGIDILISTSSMPKMLAIFWFNSTTIQFDACLLQMFAIHSLSGMESTVLLAMAFDRYVAICHPLRHATVLTLPRVTKIGVAAVVRGAALMAPLPVFIKQLPFCRSNILSHSYCLHQDVMKLACDDIRVNVVYGLIVIISAIGLDSLLISFSYLLILKTVLGLTREAQAKAFGTCVSHVCAVFIFYVPFIGLSMVHRFSKRRDSPLPVILANIYLLVPPVLNPIVYGVKTKEIRQRILRLFHVATHASEP | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.22 | 0.87 | 2.38 | Download | ---------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.22 | 0.85 | 2.03 | Download | -----------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 3 | 4iaqA | 0.19 | 0.21 | 0.85 | 1.88 | Download | --------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLP-------PFFWRQASE--------------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIAARERKATKTLGIILGAFIVCWLPFFIISLVMPI-----HLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 4 | 4ib4 | 0.16 | 0.25 | 0.88 | 1.51 | Download | -------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKG--IETN----PNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAASNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSNQTQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---- | |||||||||||||||||||
| 5 | 3uonA | 0.18 | 0.19 | 0.86 | 1.12 | Download | ----------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFI--VGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR---REKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.22 | 0.87 | 2.59 | Download | -----------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 7 | 3d4s | 0.17 | 0.19 | 0.87 | 1.72 | Download | ------------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRA----THQEAINCYA----EETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL------- | |||||||||||||||||||
| 8 | 2z73A | 0.16 | 0.24 | 0.96 | 3.03 | Download | -ETWWYNPSIVVHPHWREFDQVDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKWIGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFG-----WGAEGVLCNCSFDY-------ISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQF | |||||||||||||||||||
| 9 | 5tgzA | 0.19 | 0.22 | 0.90 | 3.02 | Download | GRGENFMDIEC--FMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 10 | 4gpoA | 0.18 | 0.21 | 0.87 | 4.56 | Download | ---------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL---------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
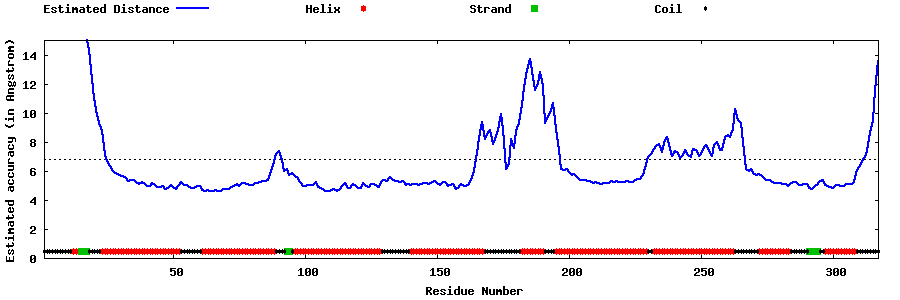
| Generated 3D models | Estimated local accuracy of models | ||
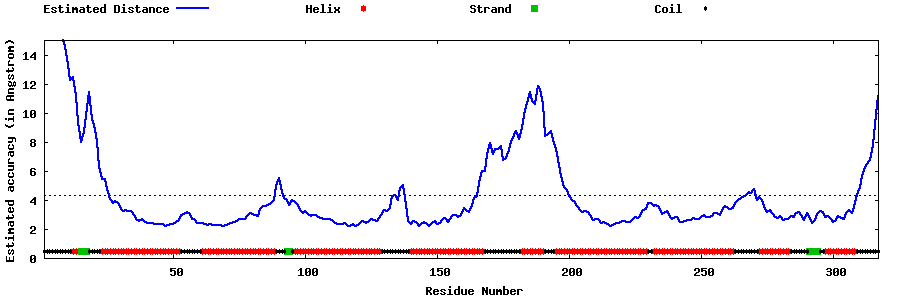 | |||
|
|
|||
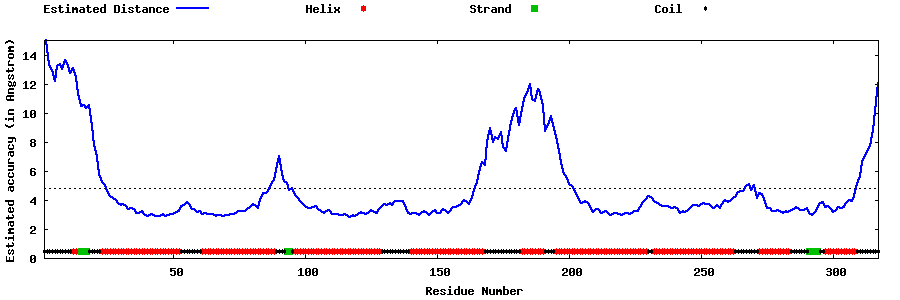 | |||
|
| |||
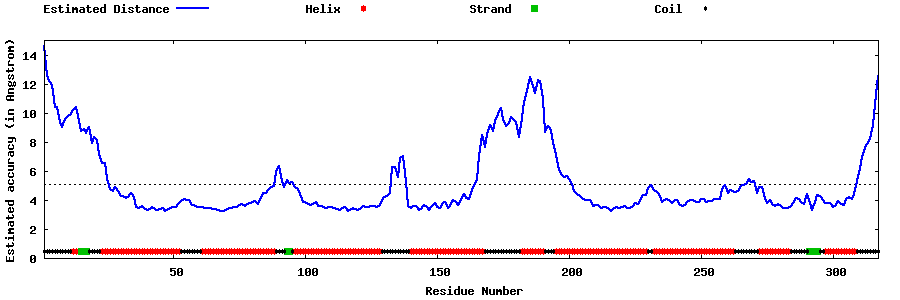 | |||
|
| |||
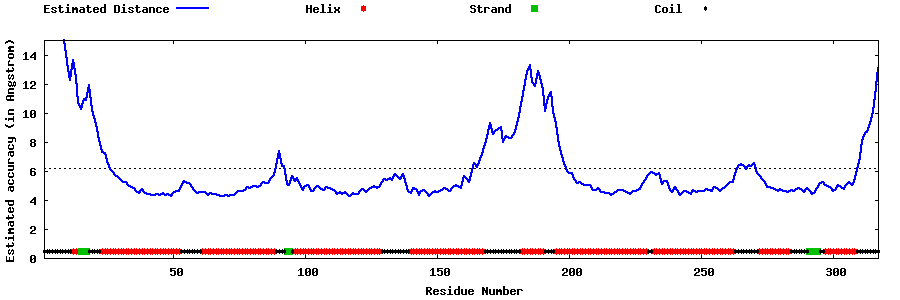 | |||
|
| |||
 | |||
|
| |||