

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MGFYHVGQAAFELLTSSFILVGFSDRPHLELIVFVVVLIFYLLTLLGNMTIVLLSALDSRLHTPMYFFLANLSFLDMCFTTGSIPQMLYNLWGPDKTISYVGCAIQLYFVLALGGVECVLLAVMAYDRYAAVCKPLHYTIIMHPRLCGQLASVAWLSGFGNSLIMAPQTLMLPRCGHRRVDHFLCEMPALIGMACVDTMMLEALAFALAIFIILAPLILILISYGYVGGTVLRIKSAAGRKKAFNTCSSHLIVVSLFYGTIIYMYLQPANTYSQDQGKFLTLFYTIVTPSVNPLIYTLRNKDVKEAMKKVLGKGSAEI | |
| CCCHHCCCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHCCCSSSSSCCCCCCCCCCCCSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCC | |
| 970212256888310356896489984169999999999999999988999788862887667578886679998788762478999998468995671899999999999999999999999976517761644457713588899999999999999999999999845489998937897148288899960475099999999999999998999999999999999308660436405316686899799975011058817899998878938986540454331246431552999999999986343789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MGFYHVGQAAFELLTSSFILVGFSDRPHLELIVFVVVLIFYLLTLLGNMTIVLLSALDSRLHTPMYFFLANLSFLDMCFTTGSIPQMLYNLWGPDKTISYVGCAIQLYFVLALGGVECVLLAVMAYDRYAAVCKPLHYTIIMHPRLCGQLASVAWLSGFGNSLIMAPQTLMLPRCGHRRVDHFLCEMPALIGMACVDTMMLEALAFALAIFIILAPLILILISYGYVGGTVLRIKSAAGRKKAFNTCSSHLIVVSLFYGTIIYMYLQPANTYSQDQGKFLTLFYTIVTPSVNPLIYTLRNKDVKEAMKKVLGKGSAEI | |
| 754343554413020210000000432400110013023313313333310010010023000000010200001001010011030000011652201000022011201300220020001002000000021020000003300100011001100310310001001020024130000000311001000210330012023313331331231022012200200010313612310110000000000011200100001041632553300000210332033003101010310130022004344564 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHCCCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHCCCSSSSSCCCCCCCCCCCCSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCC MGFYHVGQAAFELLTSSFILVGFSDRPHLELIVFVVVLIFYLLTLLGNMTIVLLSALDSRLHTPMYFFLANLSFLDMCFTTGSIPQMLYNLWGPDKTISYVGCAIQLYFVLALGGVECVLLAVMAYDRYAAVCKPLHYTIIMHPRLCGQLASVAWLSGFGNSLIMAPQTLMLPRCGHRRVDHFLCEMPALIGMACVDTMMLEALAFALAIFIILAPLILILISYGYVGGTVLRIKSAAGRKKAFNTCSSHLIVVSLFYGTIIYMYLQPANTYSQDQGKFLTLFYTIVTPSVNPLIYTLRNKDVKEAMKKVLGKGSAEI | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.23 | 0.87 | 3.50 | Download | --------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.22 | 0.89 | 2.17 | Download | -------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSM-------- | |||||||||||||||||||
| 3 | 5tgzA | 0.18 | 0.22 | 0.86 | 2.21 | Download | -----GGRGENFMD---------IENPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHR-KDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 4 | 4ib4 | 0.16 | 0.22 | 0.87 | 1.53 | Download | -----------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK--GIE----TNPNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEEQRASKVLGIVFFLFLLMWCPFFITNITLVCDSNQQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR-- | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.23 | 0.91 | 1.24 | Download | VKAEQLKTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.23 | 0.87 | 3.80 | Download | ---------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.24 | 0.82 | 1.72 | Download | ---------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF---------WRQA--SECVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLRERKATKT---LGIILGAFIVCWLPFFIISLMPIHL-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK---- | |||||||||||||||||||
| 8 | 4ea3A | 0.16 | 0.20 | 0.79 | 2.92 | Download | -------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAI--------MGSAQVEIPTPQDYWGPVFAICIFLFSF-----------------IVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLG-----VQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR---------- | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.23 | 0.87 | 4.74 | Download | --------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.23 | 0.90 | 5.56 | Download | -----------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| ||||||||||||||||||||||||||
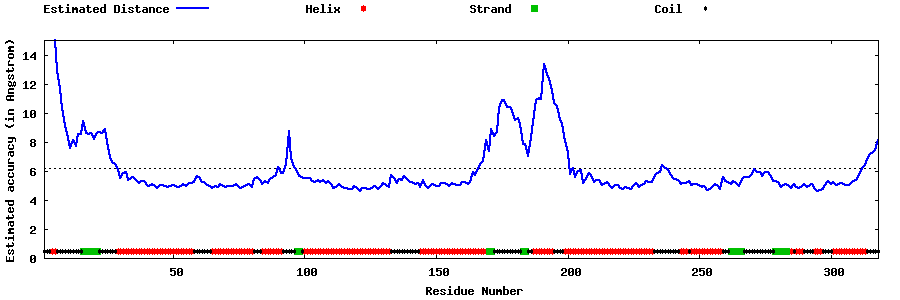
| Generated 3D models | Estimated local accuracy of models | ||
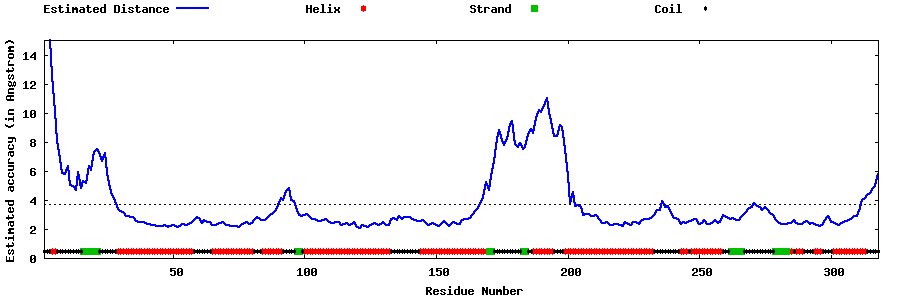 | |||
|
|
|||
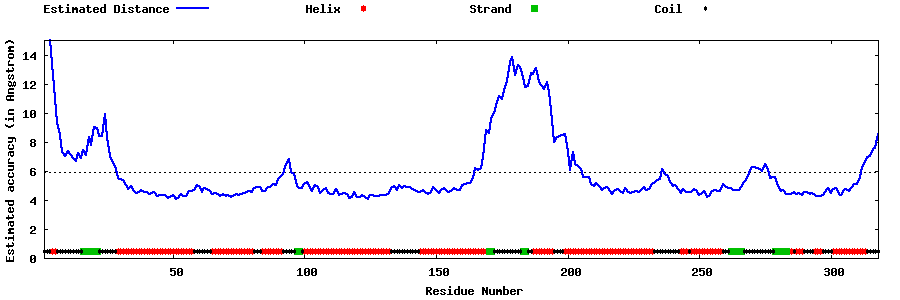 | |||
|
| |||
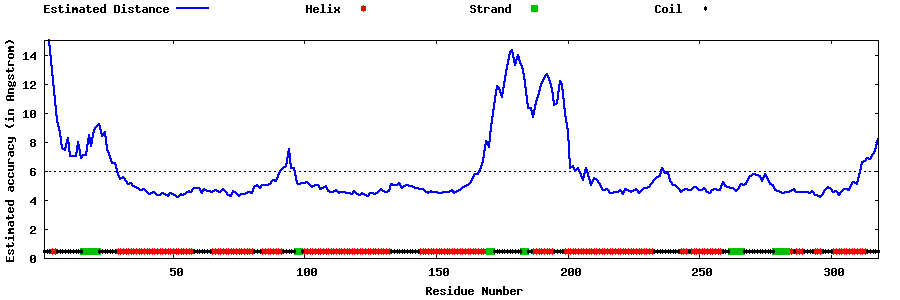 | |||
|
| |||
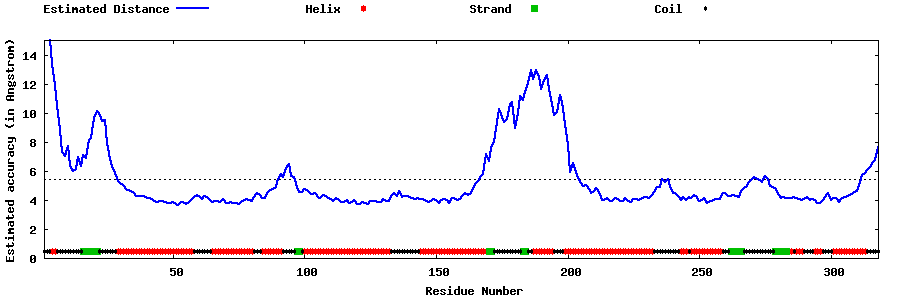 | |||
|
| |||
 | |||
|
| |||