

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MERNHNPDNCNVLNFFFADKKNKRRNFGQIVSDVGRICYSVSLSLGEPTTMGRNNLTRPSEFILLGLSSRPEDQKPLFAVFLPIYLITVIGNLLIILAIRSDTRLQTPMYFFLSILSFVDICYVTVIIPKMLVNFLSETKTISYSECLTQMYFFLAFGNTDSYLLAAMAIDRYVAICNPFHYITIMSHRCCVLLLVLSFCIPHFHSLLHILLTNQLIFCASNVIHHFFCDDQPVLKLSCSSHFVKEITVMTEGLAVIMTPFSCIIISYLRILITVLKIPSAAGKRKAFSTCGSHLTVVTLFYGSISYLYFQPLSNYTVKDQIATIIYTVLTPMLNPFIYSLRNKDMKQGLAKLMHRMKCQ | |
| CCCCCCCCCCCCCSSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCHHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHCCC | |
| 987779754300001113542000120134434544201145411034427487886055251107998832678999999999999999899999999728875664999988799874362325788887988348966868999999999999999999999999875066406642177422762899999999999999999999998613389999118916785899876265533888999999999999989999999999999961145768867322560867887999856031578487988888873799860044503355643256288999999998630088 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MERNHNPDNCNVLNFFFADKKNKRRNFGQIVSDVGRICYSVSLSLGEPTTMGRNNLTRPSEFILLGLSSRPEDQKPLFAVFLPIYLITVIGNLLIILAIRSDTRLQTPMYFFLSILSFVDICYVTVIIPKMLVNFLSETKTISYSECLTQMYFFLAFGNTDSYLLAAMAIDRYVAICNPFHYITIMSHRCCVLLLVLSFCIPHFHSLLHILLTNQLIFCASNVIHHFFCDDQPVLKLSCSSHFVKEITVMTEGLAVIMTPFSCIIISYLRILITVLKIPSAAGKRKAFSTCGSHLTVVTLFYGSISYLYFQPLSNYTVKDQIATIIYTVLTPMLNPFIYSLRNKDMKQGLAKLMHRMKCQ | |
| 764443663020031213444334434453134222112102221322440455030200100000014415212000110030122013212100100000120000000000000010111100230200000105334030300100110000002100000000001000000100100010012000000120121001001000000000100362401000001320030000002100010122133313312310230111000000102126122100000000000000111000000000013334322000000012103300200001031014003300333438 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCSSSSCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCHHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHCCC MERNHNPDNCNVLNFFFADKKNKRRNFGQIVSDVGRICYSVSLSLGEPTTMGRNNLTRPSEFILLGLSSRPEDQKPLFAVFLPIYLITVIGNLLIILAIRSDTRLQTPMYFFLSILSFVDICYVTVIIPKMLVNFLSETKTISYSECLTQMYFFLAFGNTDSYLLAAMAIDRYVAICNPFHYITIMSHRCCVLLLVLSFCIPHFHSLLHILLTNQLIFCASNVIHHFFCDDQPVLKLSCSSHFVKEITVMTEGLAVIMTPFSCIIISYLRILITVLKIPSAAGKRKAFSTCGSHLTVVTLFYGSISYLYFQPLSNYTVKDQIATIIYTVLTPMLNPFIYSLRNKDMKQGLAKLMHRMKCQ | |||||||||||||||||||||||||
| 1 | 3emlA | 0.22 | 0.21 | 0.77 | 3.21 | Download | ----------------------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.21 | 0.78 | 1.98 | Download | ---------------------------------------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-----------------GWNCEKLQSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFMLCLLNSTVNPIIYALRSKDLRHAFRSM------- | |||||||||||||||||||
| 3 | 5tgzA | 0.20 | 0.21 | 0.76 | 2.00 | Download | -------------------------------------------------GGRGENFMD---------IENPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------ | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.19 | 0.78 | 1.52 | Download | ---------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE---DVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.23 | 0.91 | 1.24 | Download | DAQKATPPKLEDKS--PDSPEMKDF--RHGFDILVGQIDAKEAQAAAEQLKTTRN-AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV-FF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVGIRIVTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------ | |||||||||||||||||||
| 6 | 3emlA | 0.22 | 0.21 | 0.76 | 3.49 | Download | -----------------------------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.18 | 0.73 | 1.71 | Download | -----------------------------------------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVNT-----------------DHIL---YTVYSTVGAFYFPTLLLIALYGRIYVEARSRIAAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH-LAIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 8 | 2rh1A | 0.18 | 0.21 | 0.73 | 2.96 | Download | ---------------------------------------------------------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQM----HWYRATHQEAINCYAEE----TCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDEGL-------------RLKIYKDTEGY-------YTIGINTNGVITKDEAEKLFNQDVDAAVRGILRNAKLK | |||||||||||||||||||
| 9 | 3emlA | 0.22 | 0.21 | 0.76 | 4.28 | Download | ---------------------------------------------------------------IM-------GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPL-MYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 10 | 3sn6R | 0.18 | 0.21 | 0.98 | 4.74 | Download | IGRNTNGVIAALINMVFQMGETGVAGFTNSLRMLRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTGTWDAEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC----- | |||||||||||||||||||
| ||||||||||||||||||||||||||
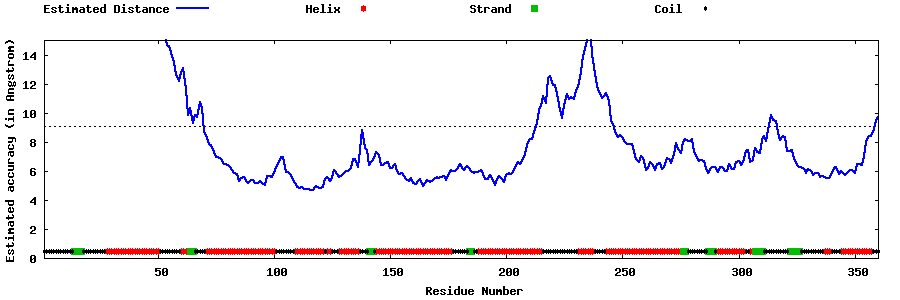
| Generated 3D models | Estimated local accuracy of models | ||
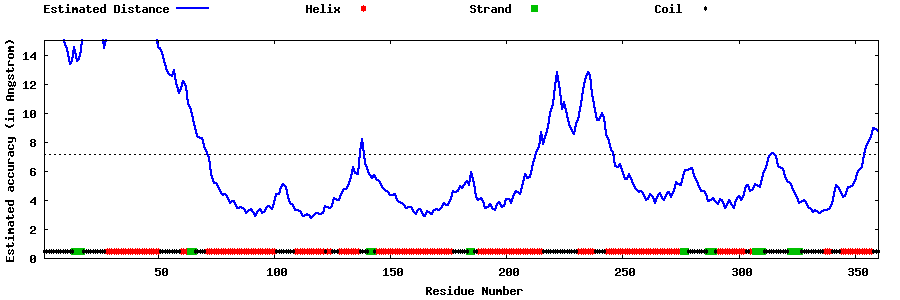 | |||
|
|
|||
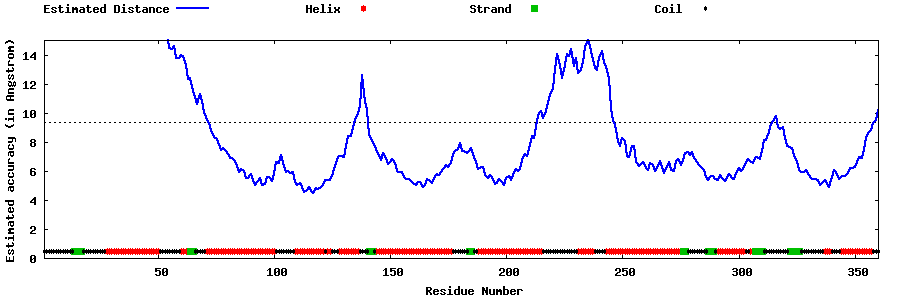 | |||
|
| |||
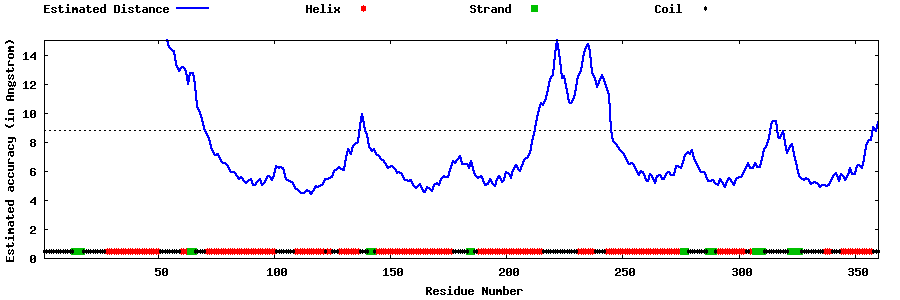 | |||
|
| |||
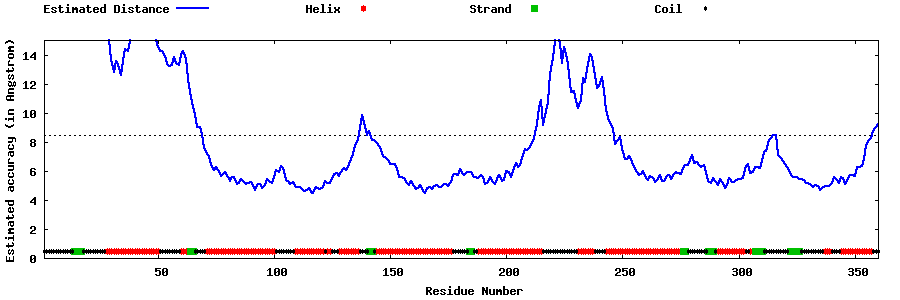 | |||
|
| |||
 | |||
|
| |||