

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGMSNLTRLSEFILLGLSSRSEDQRPLFALFLIIYLVTLMGNLLIILAIHSDPRLQNPMYFFLSILSFADICYTTVIVPKMLVNFLSEKKTISYAECLAQMYFFLVFGNIDSYLLAAMAINRCVAICNPFHYVTVMNRRCCVLLLAFPITFSYFHSLLHVLLVNRLTFCTSNVIHHFFCDVNPVLKLSCSSTFVNEIVAMTEGLASVMAPFVCIIISYLRILIAVLKIPSAAGKHKAFSTCSSHLTVVILFYGSISYVYLQPLSSYTVKDRIATINYTVLTSVLNPFIYSLRNKDMKRGLQKLINKIKSQMSRFSTKTNKICGP | |
| CCCCCCCCCHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCHHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHHCCCCCSSCCCCCCCCC | |
| 999888505646300799882256889999999999999989999999962887566499998879987436131588887698834896776899999999999999999999999987507750654207742276189999999999999999999999873238999904890678689987636553388899999999999988999999999999996004576886732256086789799985712158848798888887379986015551435664235628899999999985224135100455666798 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGMSNLTRLSEFILLGLSSRSEDQRPLFALFLIIYLVTLMGNLLIILAIHSDPRLQNPMYFFLSILSFADICYTTVIVPKMLVNFLSEKKTISYAECLAQMYFFLVFGNIDSYLLAAMAINRCVAICNPFHYVTVMNRRCCVLLLAFPITFSYFHSLLHVLLVNRLTFCTSNVIHHFFCDVNPVLKLSCSSTFVNEIVAMTEGLASVMAPFVCIIISYLRILIAVLKIPSAAGKHKAFSTCSSHLTVVILFYGSISYVYLQPLSSYTVKDRIATINYTVLTSVLNPFIYSLRNKDMKRGLQKLINKIKSQMSRFSTKTNKICGP | |
| 865723130010000001542621200011003012201332210000000012000000000000001011110022030010111534403140010011001000210000000001100000010020001001300000012002000100100000000010037240100001232003000110220101022213330331331033011100000010213613210000000000000011100200000002233432200000001310330020000203102300230033231223313344463558 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCHHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHHCCCCCSSCCCCCCCCC MGMSNLTRLSEFILLGLSSRSEDQRPLFALFLIIYLVTLMGNLLIILAIHSDPRLQNPMYFFLSILSFADICYTTVIVPKMLVNFLSEKKTISYAECLAQMYFFLVFGNIDSYLLAAMAINRCVAICNPFHYVTVMNRRCCVLLLAFPITFSYFHSLLHVLLVNRLTFCTSNVIHHFFCDVNPVLKLSCSSTFVNEIVAMTEGLASVMAPFVCIIISYLRILIAVLKIPSAAGKHKAFSTCSSHLTVVILFYGSISYVYLQPLSSYTVKDRIATINYTVLTSVLNPFIYSLRNKDMKRGLQKLINKIKSQMSRFSTKTNKICGP | |||||||||||||||||||||||||
| 1 | 3emlA | 0.23 | 0.23 | 0.85 | 3.48 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------------- | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.22 | 0.86 | 2.09 | Download | -RGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-----------------GWNCEKLQSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFMLCLLNSTVNPIIYALRSKDLRHAFRSM--------------------- | |||||||||||||||||||
| 3 | 5tgzA | 0.20 | 0.22 | 0.84 | 1.91 | Download | GRGENFMD---------IENPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.18 | 0.24 | 0.85 | 1.53 | Download | -----------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK--GIE----TNPNNITCVLTK-------E--RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLNEQRASKVLGIVFFLFLLMWCPFFITNILVLCDNTQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------- | |||||||||||||||||||
| 5 | 4yay | 0.18 | 0.21 | 0.87 | 1.24 | Download | KTTRN-AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV-FF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVGIRIVTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------------------- | |||||||||||||||||||
| 6 | 3emlA | 0.24 | 0.23 | 0.85 | 3.75 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------------- | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.20 | 0.81 | 1.71 | Download | ---------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIHL--AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF------------------ | |||||||||||||||||||
| 8 | 4ea3A | 0.18 | 0.18 | 0.77 | 2.92 | Download | -------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAIMGSAQV---------EDEEIDYWGPVFAICIFLFSF-----------------IVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLGV----QPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR----------------------- | |||||||||||||||||||
| 9 | 3emlA | 0.23 | 0.23 | 0.85 | 4.76 | Download | -------------IM-------GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPL-MYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------------- | |||||||||||||||||||
| 10 | 2ydoA | 0.19 | 0.22 | 0.92 | 5.71 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFKAAAAENLY | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
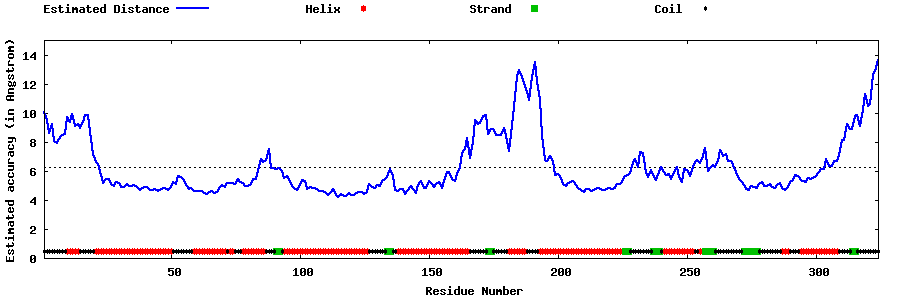 | |||
|
| |||
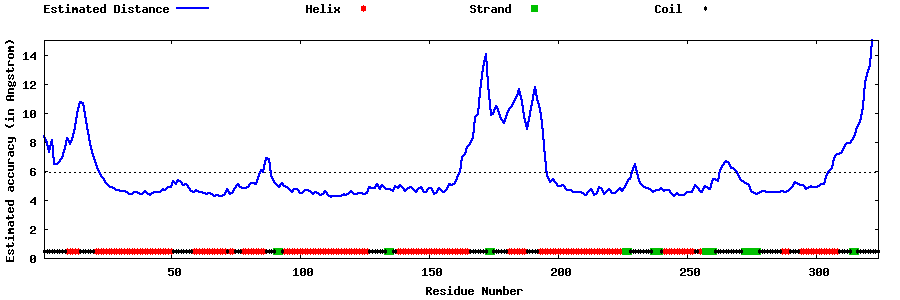 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||