

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MGRGNSTEVTEFHLLGFGVQHEFQHVLFIVLLLIYVTSLIGNIGMILLIKTDSRLQTPMYFFPQHLAFVDICYTSAITPKMLQSFTEENNLITFRGCVIQFLVYATFATSDCYLLAIMAMDCYVAICKPLRYPMIMSQTVYIQLVAGSYIIGSINASVHTGFTFSLSFCKSNKINHFFCDGLPILALSCSNIDINIILDVVFVGFDLMFTELVIIFSYIYIMVTILKMSSTAGRKKSFSTCASHLTAVTIFYGTLSYMYLQPQSNNSQENMKVASIFYGTVIPMLNPLIYSLRNKEGK | |
| CCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCCCCC | |
| 9998873231357864789936089999999999999998886999998518887773888876799884472114609999987248978858999999999999999999999999754134016220288016788999999999999999999999998454078999148541581888877056862667778888899999999999999999999980367757675432255889997999974322458378999878789789988211141243563324555689 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MGRGNSTEVTEFHLLGFGVQHEFQHVLFIVLLLIYVTSLIGNIGMILLIKTDSRLQTPMYFFPQHLAFVDICYTSAITPKMLQSFTEENNLITFRGCVIQFLVYATFATSDCYLLAIMAMDCYVAICKPLRYPMIMSQTVYIQLVAGSYIIGSINASVHTGFTFSLSFCKSNKINHFFCDGLPILALSCSNIDINIILDVVFVGFDLMFTELVIIFSYIYIMVTILKMSSTAGRKKSFSTCASHLTAVTIFYGTLSYMYLQPQSNNSQENMKVASIFYGTVIPMLNPLIYSLRNKEGK | |
| 8565230300100000004325010000130332132123233101200200130000001002000010010000000300020025433010300020110000000000100000001000000211202040033000000220101003001010000030201432200000003200030001323110110021013223313310330331011000103126323101000110310010232012000010324313344200002103300230030001224638 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHCCCCCCCC MGRGNSTEVTEFHLLGFGVQHEFQHVLFIVLLLIYVTSLIGNIGMILLIKTDSRLQTPMYFFPQHLAFVDICYTSAITPKMLQSFTEENNLITFRGCVIQFLVYATFATSDCYLLAIMAMDCYVAICKPLRYPMIMSQTVYIQLVAGSYIIGSINASVHTGFTFSLSFCKSNKINHFFCDGLPILALSCSNIDINIILDVVFVGFDLMFTELVIIFSYIYIMVTILKMSSTAGRKKSFSTCASHLTAVTIFYGTLSYMYLQPQSNNSQENMKVASIFYGTVIPMLNPLIYSLRNKEGK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.20 | 0.88 | 3.53 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFR | |||||||||||||||||||
| 2 | 5tgzA | 0.18 | 0.21 | 0.90 | 1.90 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLR | |||||||||||||||||||
| 3 | 5tjvA | 0.15 | 0.22 | 0.90 | 2.00 | Download | -------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHV-FHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCESDIFPHID----------------------ETYLMFWIGVTSVLLLFIVYAYMYILWKARAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLR | |||||||||||||||||||
| 4 | 3uon | 0.17 | 0.25 | 0.90 | 1.56 | Download | --------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQF---------FSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCCIPNTVWTIGYWLCYINSTINPACYALCNATFK | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.24 | 0.93 | 1.28 | Download | KTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHR-NVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFK | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.20 | 0.88 | 3.76 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFR | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.20 | 0.62 | 1.68 | Download | ---------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSR---------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4ea3A | 0.16 | 0.20 | 0.86 | 2.34 | Download | -------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEIECLVEIPTPQDYWGPVFAICIFLFSF-----------------IVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLG-----VQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFK | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.20 | 0.88 | 5.14 | Download | ----------------------ISSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFR | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.20 | 0.92 | 5.97 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFR | |||||||||||||||||||
| ||||||||||||||||||||||||||
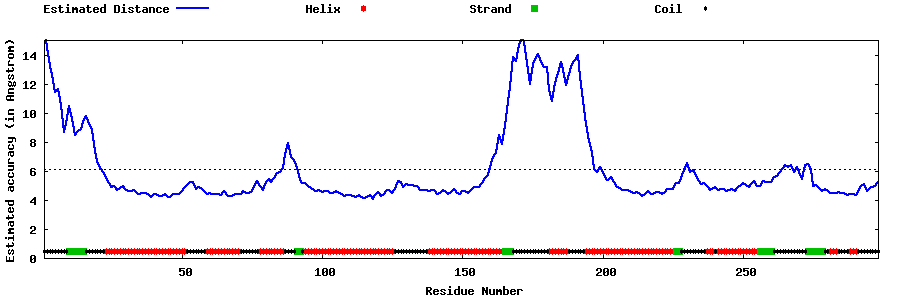
| Generated 3D models | Estimated local accuracy of models | ||
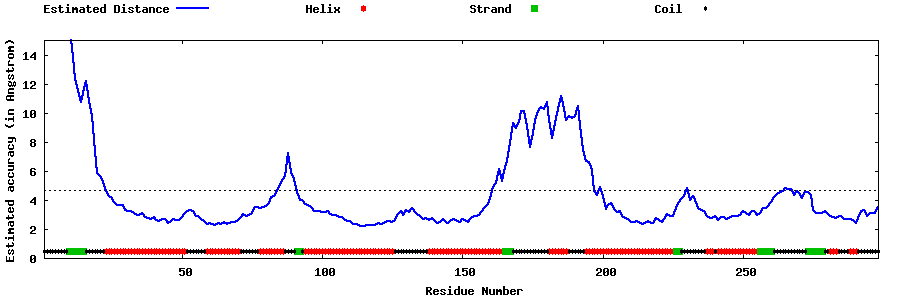 | |||
|
|
|||
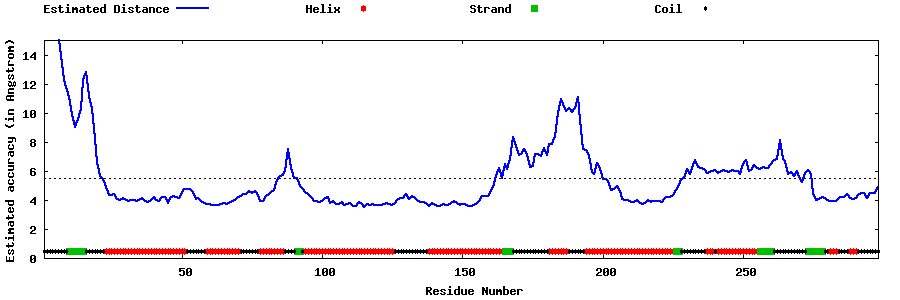 | |||
|
| |||
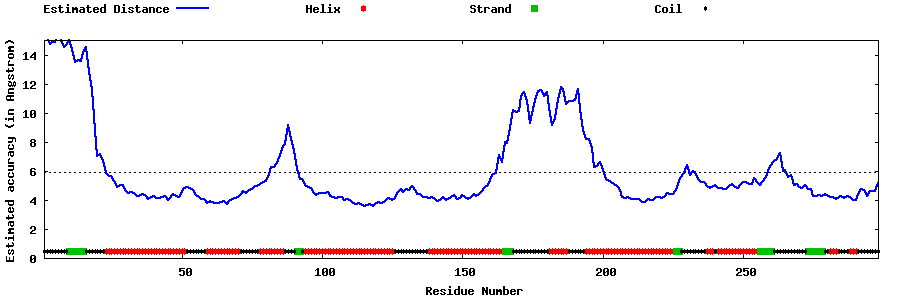 | |||
|
| |||
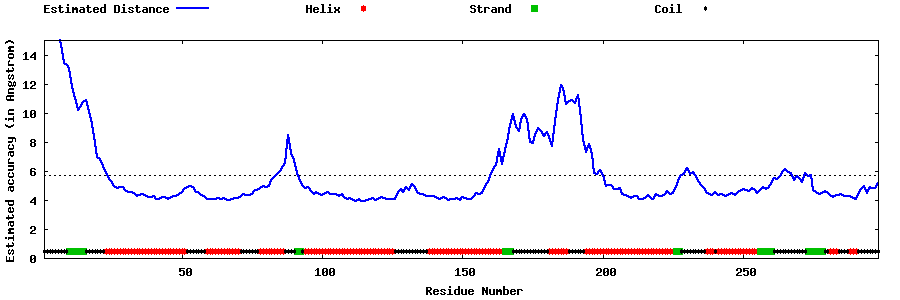 | |||
|
| |||
 | |||
|
| |||