

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MLEGVEHLLLLLLLTDVNSKELQSGNQTSVSHFILVGLHHPPQLGAPLFLAFLVIYLLTVSGNGLIILTVLVDIRLHRPMCLFLCHLSFLDMTISCAIVPKMLAGFLLGSRIISFGGCVIQLFSFHFLGCTECFLYTLMAYDRFLAICKPLHYATIMTHRVCNSLALGTWLGGTIHSLFQTSFVFRLPFCGPNRVDYIFCDIPAMLRLACADTAINELVTFADIGFLALTCFMLILTSYGYIVAAILRIPSADGRRNAFSTCAAHLTVVIVYYVPCTFIYLRPCSQEPLDGVVAVFYTVITPLLNSIIYTLCNKEMKAALQRLGGHKEVQPH | |
| CCHHHHHHHHHHHHCCCCHHHHHCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCHHHHHHHHHHHHHCCHHHHHCCCCHHHHHHHHHHHHCCCCCCC | |
| 91057767898861555663552566871157889468998023899999999999999987888566343069989978999887998865234346499999860699677289999999999999999999999998651776373314700168759999999999999999999999963689989885777613818888885314047788999998899999999999999999999601757434248988788799907456206568882889998504556888854664101274323519999999999961567999 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MLEGVEHLLLLLLLTDVNSKELQSGNQTSVSHFILVGLHHPPQLGAPLFLAFLVIYLLTVSGNGLIILTVLVDIRLHRPMCLFLCHLSFLDMTISCAIVPKMLAGFLLGSRIISFGGCVIQLFSFHFLGCTECFLYTLMAYDRFLAICKPLHYATIMTHRVCNSLALGTWLGGTIHSLFQTSFVFRLPFCGPNRVDYIFCDIPAMLRLACADTAINELVTFADIGFLALTCFMLILTSYGYIVAAILRIPSADGRRNAFSTCAAHLTVVIVYYVPCTFIYLRPCSQEPLDGVVAVFYTVITPLLNSIIYTLCNKEMKAALQRLGGHKEVQPH | |
| 73441431000001232436414452313000000000033450010001312331331333232000002004301000010023000100110010002000000164210004000000011022011002000100300000002101000000330000001101300120030013101302000113000000012000300010033101311230133033213303311210010001321461132001001001000201311000000003232310000003102310331030000205202400330154343378 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHHHHHHHHHHCCCCHHHHHCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCHHHHHHHHHHHHHCCHHHHHCCCCHHHHHHHHHHHHCCCCCCC MLEGVEHLLLLLLLTDVNSKELQSGNQTSVSHFILVGLHHPPQLGAPLFLAFLVIYLLTVSGNGLIILTVLVDIRLHRPMCLFLCHLSFLDMTISCAIVPKMLAGFLLGSRIISFGGCVIQLFSFHFLGCTECFLYTLMAYDRFLAICKPLHYATIMTHRVCNSLALGTWLGGTIHSLFQTSFVFRLPFCGPNRVDYIFCDIPAMLRLACADTAINELVTFADIGFLALTCFMLILTSYGYIVAAILRIPSADGRRNAFSTCAAHLTVVIVYYVPCTFIYLRPCSQEPLDGVVAVFYTVITPLLNSIIYTLCNKEMKAALQRLGGHKEVQPH | |||||||||||||||||||||||||
| 1 | 3emlA | 0.17 | 0.21 | 0.83 | 3.26 | Download | -----------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHII-NCFTFFCPDCSHLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.21 | 0.84 | 2.33 | Download | ----------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL---------QSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM--------- | |||||||||||||||||||
| 3 | 4iaqA | 0.19 | 0.22 | 0.77 | 2.12 | Download | ---------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNT-------------------YTVYSTVGAF-----YFPTLLLIALYGRIYVEARSQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.22 | 0.84 | 1.50 | Download | ----------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE---DVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------ | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.22 | 0.91 | 1.22 | Download | LVGQIDDALKLANEGKVKAEQLKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKA-LK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGRVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------- | |||||||||||||||||||
| 6 | 3emlA | 0.17 | 0.21 | 0.83 | 3.36 | Download | ------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHII-NCFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 7 | 4iaq | 0.22 | 0.21 | 0.79 | 1.73 | Download | ------------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 8 | 4buoA | 0.14 | 0.19 | 0.86 | 2.92 | Download | -------------------------NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGPIVDLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYI--------SDEQWFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL-------- | |||||||||||||||||||
| 9 | 5tgzA | 0.19 | 0.21 | 0.84 | 4.25 | Download | --------------------GGRGENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 10 | 2ydoA | 0.15 | 0.20 | 0.86 | 5.08 | Download | --------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQ | |||||||||||||||||||
| ||||||||||||||||||||||||||
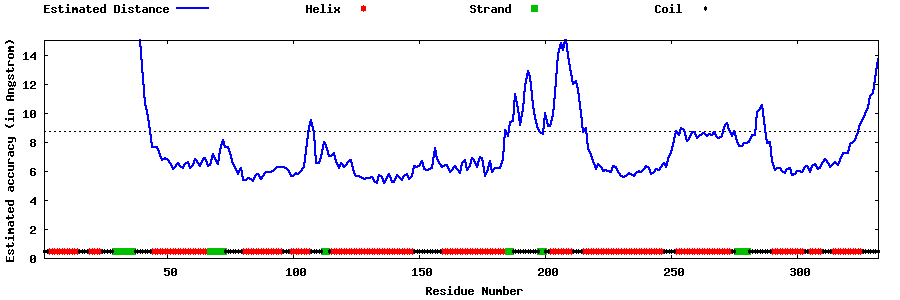
| Generated 3D models | Estimated local accuracy of models | ||
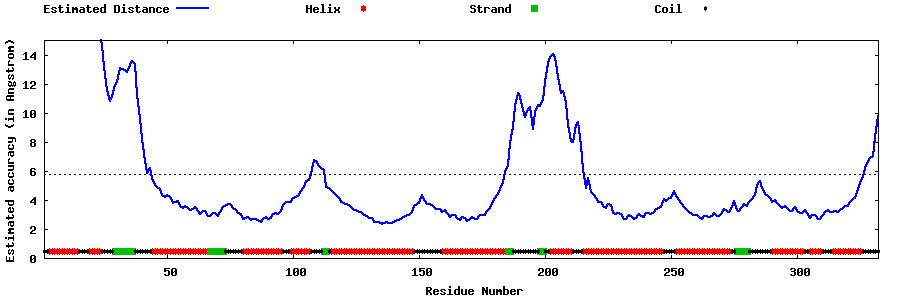 | |||
|
|
|||
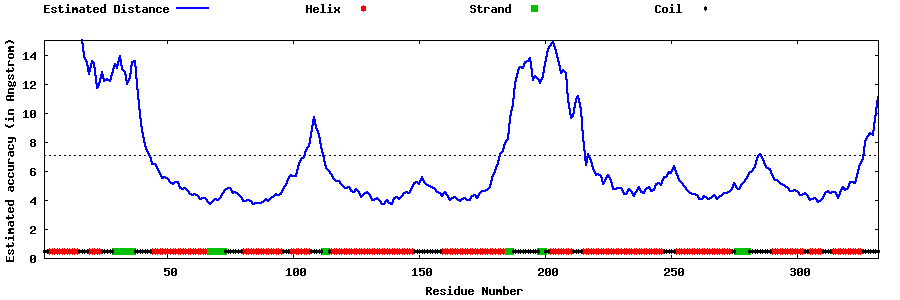 | |||
|
| |||
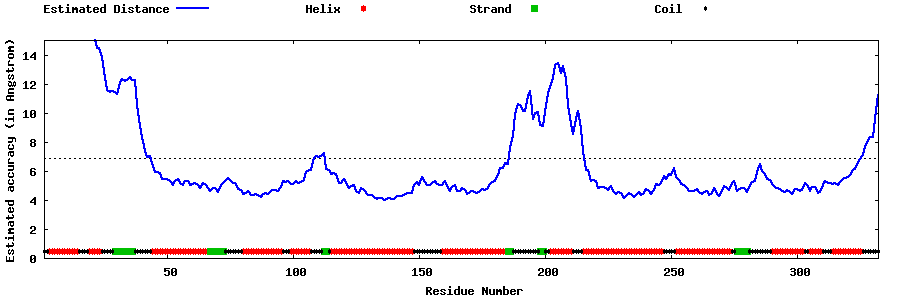 | |||
|
| |||
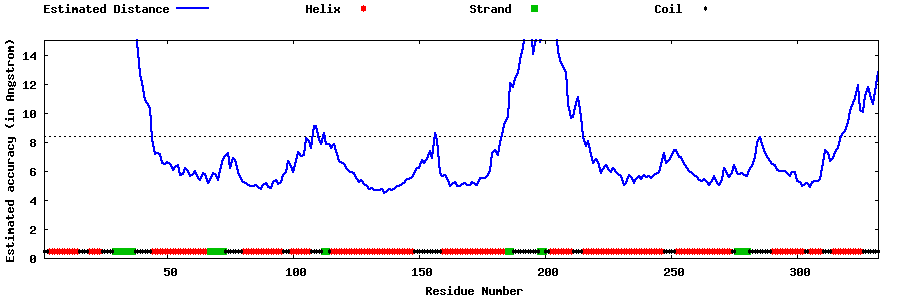 | |||
|
| |||
 | |||
|
| |||