

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDTSTSVTYDSSLQISQFILMGLPGIHEWQHWLSLPLTLLYLLALGANLLIIITIQHETVLHEPMYHLLGILAVVDIGLATTIMPKILAIFWFDAKAISLPMCFAQIYAIHCFFCIESGIFLCMAVDRYIAICRPLQYPSIVTKAFVFKATGFIMLRNGLLTIPVPILAAQRHYCSRNEIEHCLCSNLGVISLACDDITVNKFYQLMLAWVLVGSDMALVFSSYAVILHSVLRLNSAEAMSKALSTCSSHLILILFHTGIIVLSVTHLAEKKIPLIPVFLNVLHNVIPPALNPLACALRMHKLRLGFQRLLGLGQDVSK | |
| CCCCCCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHCCCCCCC | |
| 9886778999987560003679999878899999999999999999999988877468875433899999999999999985399999999469987888889999999999999999999999813167406654575118889999999999999999999899999428999989378744330767888347817989999999999999999999999999999981688976888986505989999999987888751046888995599999999963775636812216579999999999622664579 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDTSTSVTYDSSLQISQFILMGLPGIHEWQHWLSLPLTLLYLLALGANLLIIITIQHETVLHEPMYHLLGILAVVDIGLATTIMPKILAIFWFDAKAISLPMCFAQIYAIHCFFCIESGIFLCMAVDRYIAICRPLQYPSIVTKAFVFKATGFIMLRNGLLTIPVPILAAQRHYCSRNEIEHCLCSNLGVISLACDDITVNKFYQLMLAWVLVGSDMALVFSSYAVILHSVLRLNSAEAMSKALSTCSSHLILILFHTGIIVLSVTHLAEKKIPLIPVFLNVLHNVIPPALNPLACALRMHKLRLGFQRLLGLGQDVSK | |
| 6454144313242312102000001213201000110120122033112100000201420010002000100100100020010200000002045030400000012002102201000000020000000210100000023100100000032120112011010210310452000000010210020002303001110120033233312310320111002000301256013201200100100010232132001001134411000012012100300330030000203400410030024446368 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHCCCCCCC MDTSTSVTYDSSLQISQFILMGLPGIHEWQHWLSLPLTLLYLLALGANLLIIITIQHETVLHEPMYHLLGILAVVDIGLATTIMPKILAIFWFDAKAISLPMCFAQIYAIHCFFCIESGIFLCMAVDRYIAICRPLQYPSIVTKAFVFKATGFIMLRNGLLTIPVPILAAQRHYCSRNEIEHCLCSNLGVISLACDDITVNKFYQLMLAWVLVGSDMALVFSSYAVILHSVLRLNSAEAMSKALSTCSSHLILILFHTGIIVLSVTHLAEKKIPLIPVFLNVLHNVIPPALNPLACALRMHKLRLGFQRLLGLGQDVSK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.17 | 0.23 | 0.87 | 2.62 | Download | -------------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC-PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIR-SHVLRQ | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.26 | 0.84 | 2.15 | Download | --------------------------PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVIICWGPLLAIMVYDVFGKMNKLIKTVFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 3 | 4iaqA | 0.17 | 0.22 | 0.85 | 1.92 | Download | ------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTG--RWTLVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPP------FFWRQASE---------------CVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIMAARERKATKTLGIILGAFIVCWLPFFIISLVM---PIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 4 | 4ib4 | 0.20 | 0.26 | 0.87 | 1.53 | Download | -----------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKG--IET----NPNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADSNEQRASKVLGIVFFLFLMWCPFFITNITLVLCDSNQQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--- | |||||||||||||||||||
| 5 | 3uonA | 0.16 | 0.18 | 0.85 | 1.12 | Download | --------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQF--IVGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFITWAPYNVMVLINTFCAPIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------- | |||||||||||||||||||
| 6 | 3emlA | 0.16 | 0.23 | 0.87 | 2.83 | Download | ---------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.23 | 0.58 | 1.71 | Download | ---------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARS-------------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.16 | 0.18 | 0.91 | 2.98 | Download | ---------NSDL--------DVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGIVDTATLKVV----IQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL-------- | |||||||||||||||||||
| 9 | 3emlA | 0.17 | 0.23 | 0.86 | 3.35 | Download | -------------------------I--GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISF----CAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVV------------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFCPDC-SHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIR-SHVLRQ | |||||||||||||||||||
| 10 | 4gpoA | 0.17 | 0.20 | 0.87 | 4.41 | Download | -------------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCLPFFLVNIVNVFNRDVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL--------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
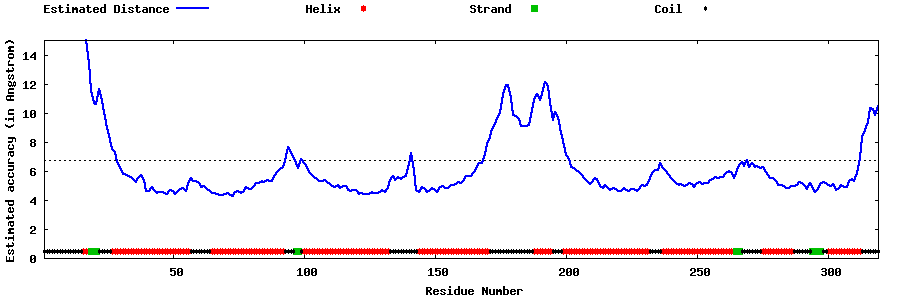
| Generated 3D models | Estimated local accuracy of models | ||
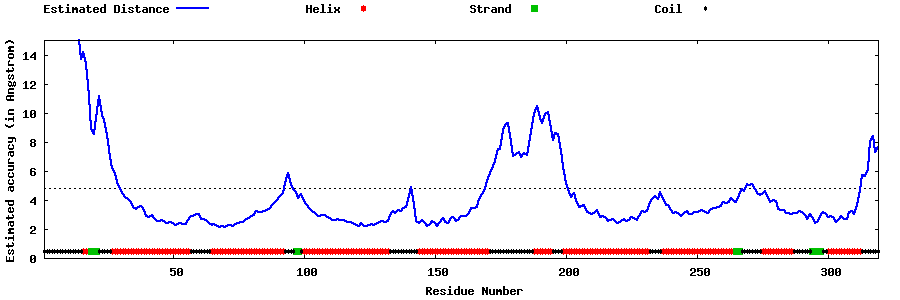 | |||
|
|
|||
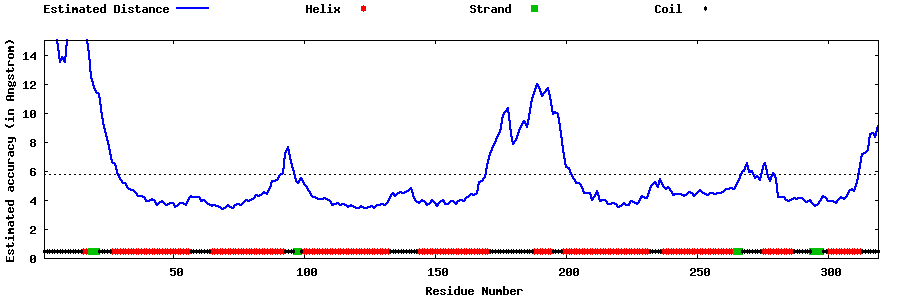 | |||
|
| |||
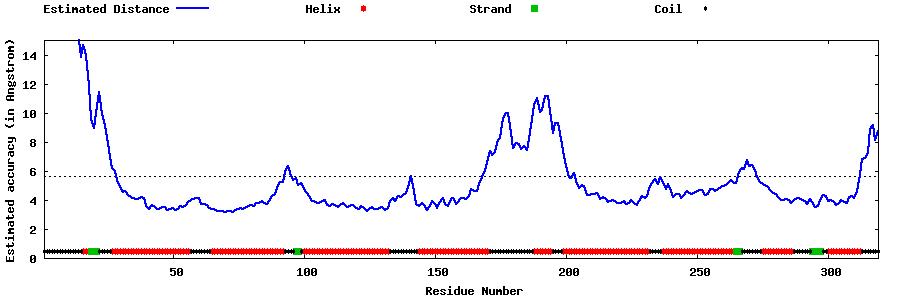 | |||
|
| |||
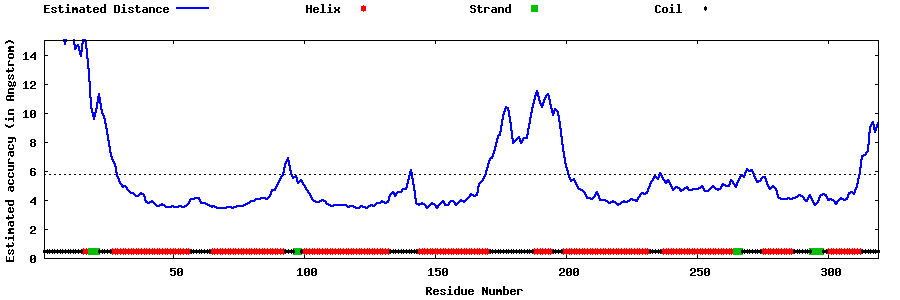 | |||
|
| |||
 | |||
|
| |||