

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDLKNGSLVTEFILLGFFGRWELQIFFFVTFSLIYGATVMGNILIMVTVTCRSTLHSPLYFLLGNLSFLDMCLSTATTPKMIIDLLTDHKTISVWGCVTQMFFMHFFGGAEMTLLIIMAFDRYVAICKPLHYRTIMSHKLLKGFAILSWIIGFLHSISQIVLTMNLPFCGHNVINNIFCDLPLVIKLACIETYTLELFVIADSGLLSFTCFILLLVSYIVILVSVPKKSSHGLSKALSTLSAHIIVVTLFFGPCIFIYVWPFSSLASNKTLAVFYTVITPLLNPSIYTLRNKKMQEAIRKLRFQYVSSAQNF | |
| CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCCCSSHHHHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHCCCCCCC | |
| 998779711477883699883168999999999999999878872562431799999789998859998676432363999998715996785899999999999999999999999986618874623035321687599999999999999999999999544899898836676318288888940340465756877767999999999999999999997310731231999888879992436630630378488999986403412564344400137433563999999999998446775579 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MDLKNGSLVTEFILLGFFGRWELQIFFFVTFSLIYGATVMGNILIMVTVTCRSTLHSPLYFLLGNLSFLDMCLSTATTPKMIIDLLTDHKTISVWGCVTQMFFMHFFGGAEMTLLIIMAFDRYVAICKPLHYRTIMSHKLLKGFAILSWIIGFLHSISQIVLTMNLPFCGHNVINNIFCDLPLVIKLACIETYTLELFVIADSGLLSFTCFILLLVSYIVILVSVPKKSSHGLSKALSTLSAHIIVVTLFFGPCIFIYVWPFSSLASNKTLAVFYTVITPLLNPSIYTLRNKKMQEAIRKLRFQYVSSAQNF | |
| 874644230000000000424400100013133323323332330000010043030000100230012001100000020000003532100050000000110331131020002003100000022110100003310000012013102301300131023030000122000100221003000020230103002301310231033033112100200132146122200100300210030132101000000333233011100310231033113000120540230033014320334763 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCCCSSHHHHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHCCCCCCC MDLKNGSLVTEFILLGFFGRWELQIFFFVTFSLIYGATVMGNILIMVTVTCRSTLHSPLYFLLGNLSFLDMCLSTATTPKMIIDLLTDHKTISVWGCVTQMFFMHFFGGAEMTLLIIMAFDRYVAICKPLHYRTIMSHKLLKGFAILSWIIGFLHSISQIVLTMNLPFCGHNVINNIFCDLPLVIKLACIETYTLELFVIADSGLLSFTCFILLLVSYIVILVSVPKKSSHGLSKALSTLSAHIIVVTLFFGPCIFIYVWPFSSLASNKTLAVFYTVITPLLNPSIYTLRNKKMQEAIRKLRFQYVSSAQNF | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.20 | 0.88 | 3.32 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYFN-----------FFACVLVPLLLMLGVYLRIFLAARRQVHAAKSLAIIVGLFALCWLPLH-IINCFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.27 | 0.89 | 2.16 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL---------QSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM----------- | |||||||||||||||||||
| 3 | 5tgzA | 0.16 | 0.27 | 0.89 | 2.07 | Download | ---GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVC-------------------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMDIELAKTLVLILVVLIICWGPLLAIKMNLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.24 | 0.88 | 1.53 | Download | -----------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIE-T-----NPNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCNQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.23 | 0.90 | 1.21 | Download | LKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------- | |||||||||||||||||||
| 6 | 5tgzA | 0.20 | 0.27 | 0.90 | 3.51 | Download | GRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV--------LPLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 7 | 4iaq | 0.21 | 0.26 | 0.84 | 1.73 | Download | --------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVNT--------------------DHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADRERKATKTLGIILGAFIVCWLPFFIISLVMPIH-LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 8 | 4buoA | 0.16 | 0.23 | 0.88 | 2.75 | Download | ----NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPI------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQAVVIAFVVCWLPYHVRRLMFCYI-----------SDEQWTTNALVYVSAAINPILYNLVSANFRQVFLSTL---------- | |||||||||||||||||||
| 9 | 5tgzA | 0.19 | 0.27 | 0.89 | 4.63 | Download | GRGENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-FAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.23 | 0.92 | 5.44 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQKQMESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEF | |||||||||||||||||||
| ||||||||||||||||||||||||||
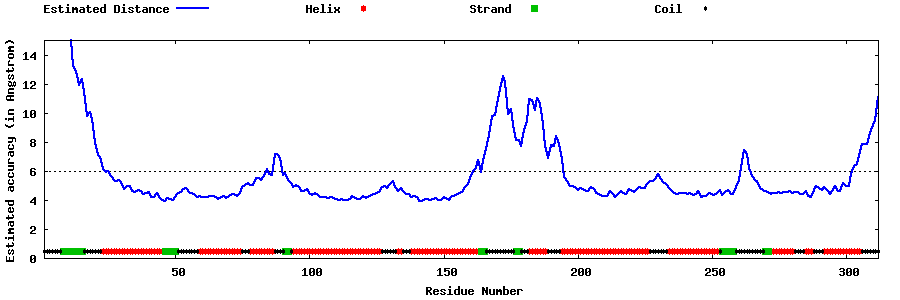
| Generated 3D models | Estimated local accuracy of models | ||
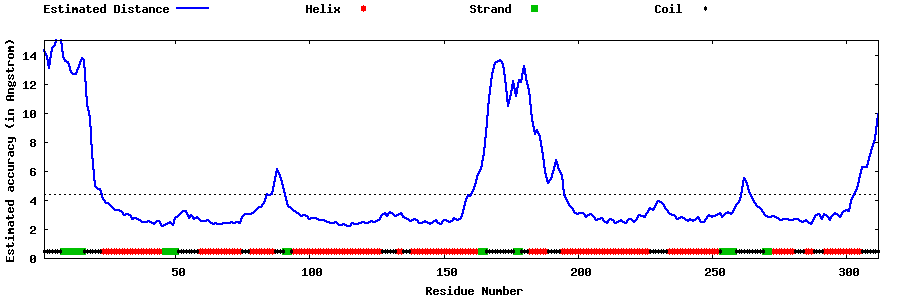 | |||
|
|
|||
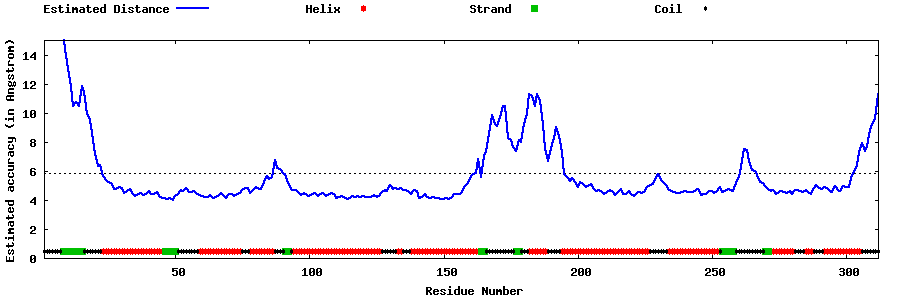 | |||
|
| |||
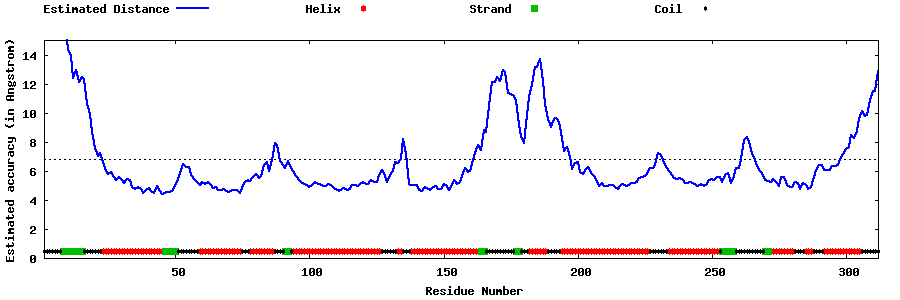 | |||
|
| |||
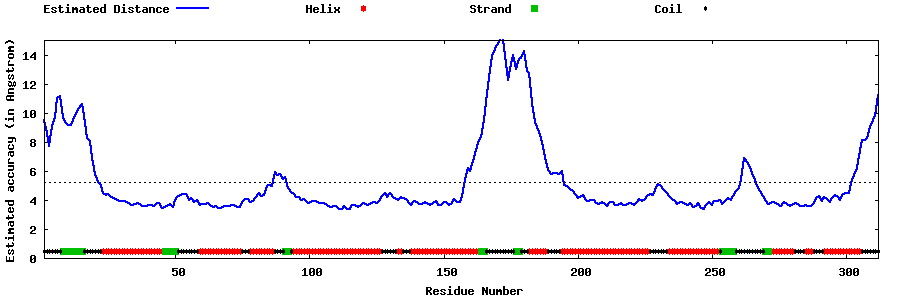 | |||
|
| |||
 | |||
|
| |||