

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MLTSLTDLCFSPIQVAEIKSLPKSMNETNHSRVTEFVLLGLSSSRELQPFLFLTFSLLYLAILLGNFLIILTVTSDSRLHTPMYFLLANLSFIDVCVASFATPKMIADFLVERKTISFDACLAQIFFVHLFTGSEMVLLVSMAYDRYVAICKPLHYMTVMSRRVCVVLVLISWFVGFIHTTSQLAFTVNLPFCGPNKVDSFFCDLPLVTKLACIDTYVVSLLIVADSGFLSLSSFLLLVVSYTVILVTVRNRSSASMAKARSTLTAHITVVTLFFGPCIFIYVWPFSSYSVDKVLAVFYTIFTLILNPVIYTLRNKEVKAAMSKLKSRYLKPSQVSVVIRNVLFLETK | |
| CCHHHHHHHCCHHHHCCCCCHHHHHHCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCHHHHHHHHHHHHHHCCCHHHCCCCHHHHHHHHHHHHHCCCHHHHHHHHHHHCCCCCC | |
| 922465453040221123534435533458600477881699983158999999999999999877874563431799999789998749987651104573899998743995784899999999999999999999999986517763631026003677499999999999999999999999240899898847776328488787830340545556888878999999999999999999996203722140899898889993536620543688588999962243378876235401037432462999999999998207832321556542045789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MLTSLTDLCFSPIQVAEIKSLPKSMNETNHSRVTEFVLLGLSSSRELQPFLFLTFSLLYLAILLGNFLIILTVTSDSRLHTPMYFLLANLSFIDVCVASFATPKMIADFLVERKTISFDACLAQIFFVHLFTGSEMVLLVSMAYDRYVAICKPLHYMTVMSRRVCVVLVLISWFVGFIHTTSQLAFTVNLPFCGPNKVDSFFCDLPLVTKLACIDTYVVSLLIVADSGFLSLSSFLLLVVSYTVILVTVRNRSSASMAKARSTLTAHITVVTLFFGPCIFIYVWPFSSYSVDKVLAVFYTIFTLILNPVIYTLRNKEVKAAMSKLKSRYLKPSQVSVVIRNVLFLETK | |
| 624212410012132362432364045503120000000000424400100003133323313331330000010033020000000230011001100000020000002532000050000000110231131010001003000000021110100003310000002013102201300130013030000122000000221002000020230003001201310221033033112000100132046122100100200200030121101000000332133010000300231033113000010540230023004330326302100221121328 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCHHHHHHHCCHHHHCCCCCHHHHHHCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCHHHHHHHHHHHHHHCCCHHHCCCCHHHHHHHHHHHHHCCCHHHHHHHHHHHCCCCCC MLTSLTDLCFSPIQVAEIKSLPKSMNETNHSRVTEFVLLGLSSSRELQPFLFLTFSLLYLAILLGNFLIILTVTSDSRLHTPMYFLLANLSFIDVCVASFATPKMIADFLVERKTISFDACLAQIFFVHLFTGSEMVLLVSMAYDRYVAICKPLHYMTVMSRRVCVVLVLISWFVGFIHTTSQLAFTVNLPFCGPNKVDSFFCDLPLVTKLACIDTYVVSLLIVADSGFLSLSSFLLLVVSYTVILVTVRNRSSASMAKARSTLTAHITVVTLFFGPCIFIYVWPFSSYSVDKVLAVFYTIFTLILNPVIYTLRNKEVKAAMSKLKSRYLKPSQVSVVIRNVLFLETK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.22 | 0.22 | 0.79 | 3.03 | Download | --------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYFN-----------FFACVLVPLLLMLGVYLRIFLAARQLVHAAKSLAIIVGLFALCWLPLHII-NCFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--------------- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.22 | 0.80 | 2.12 | Download | -------------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL---------QSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM----------------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.22 | 0.22 | 0.72 | 1.92 | Download | ------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISILYTVYSTVGAF---------------------------------------YFPTLLLIALYGRIYVEARSKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------- | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.21 | 0.80 | 1.50 | Download | -------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSLSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------------------- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.22 | 0.88 | 1.22 | Download | TPPKLSPMKDFRHGEGKAQAAAEQLKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------------------- | |||||||||||||||||||
| 6 | 3emlA | 0.22 | 0.22 | 0.79 | 3.14 | Download | ---------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARQLVHAAKSLAIIVGLFALCWLPLHII-NCFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--------------- | |||||||||||||||||||
| 7 | 4iaq | 0.23 | 0.23 | 0.76 | 1.73 | Download | --------------------------------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASECVVNT-----------------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADRERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.18 | 0.21 | 0.79 | 2.96 | Download | ----------------------------NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGTPI----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQAVVIAFVVCWLPYHVRRLMFCYI-------SDEQWTFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL---------------------- | |||||||||||||||||||
| 9 | 5tgzA | 0.19 | 0.22 | 0.80 | 4.15 | Download | -----------------------GGRGENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVYDVFGKMTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------------------- | |||||||||||||||||||
| 10 | 2ydoA | 0.20 | 0.21 | 0.85 | 4.75 | Download | -----------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLQMESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVQEPFKAAAAENLYF---- | |||||||||||||||||||
| ||||||||||||||||||||||||||
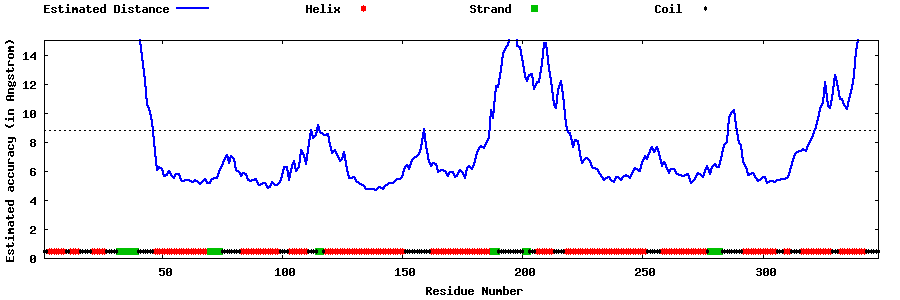
| Generated 3D models | Estimated local accuracy of models | ||
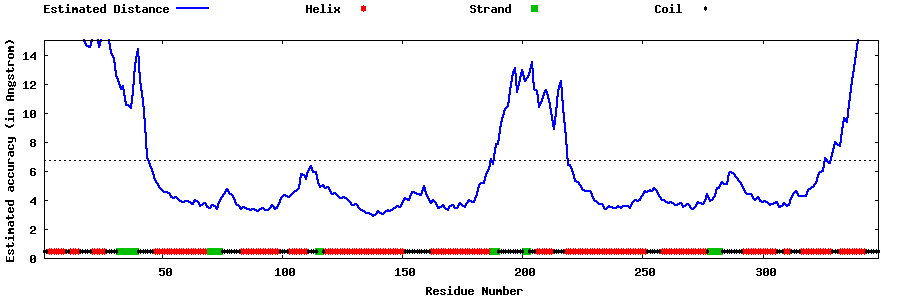 | |||
|
|
|||
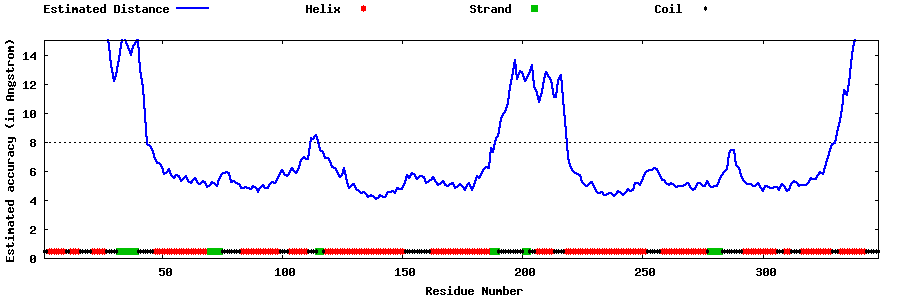 | |||
|
| |||
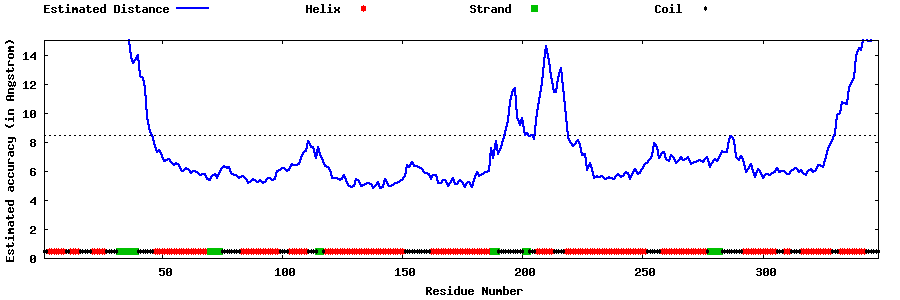 | |||
|
| |||
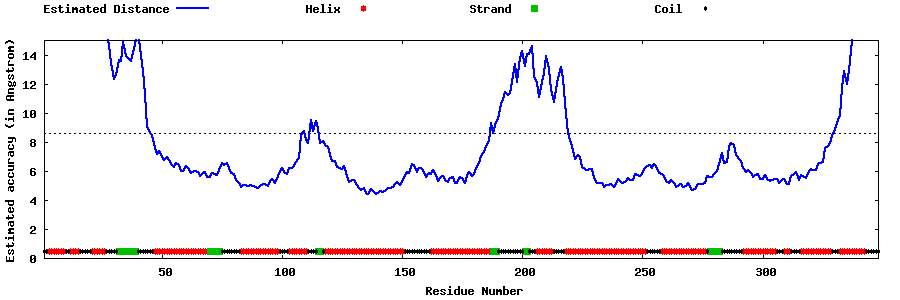 | |||
|
| |||
 | |||
|
| |||