

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGLTQDFFPPTSELLEGGNQTSTFEFLLWGLSDQPQQQHIFFLLFLWMYVVTVAGNLLIVLAIGTDTHLHTPMYFFLASLSCADIFSTSTTVPKALVNIQTQSRSISYAGCLAQLYFFLTFGDMDIFLPATMAYDRYVAICHLLHYMMIMSLHRCAFLVTACWTLTSLLAMTRTFLIFRLSLCSAILPGFFCDLGPLMKVSCSDAQVNELVLLFLGGAVILIPFMLILVSYIRIVSAILRAPSAQGRRKAFSTCDSHLVVVALFFGTVIRAYLCPSSSSSNSVKEDTAAAVMYTVVTPLLNPFIYSMRNKDMKAAVVRLLKGRVSFSQGQ | |
| CCCCCCCCCCHHHHCCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHHSSSCCCCCCCCCCCCCSSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHCCCCCCCCC | |
| 987511152111015376785065352116999821558999999999999999899999999618875552999988799875543404889988998438965758999999999999999999999999875166406643376313760899999999999999999999998511356897778167869987551643168899999999999998799999999999998500467788675205717789989998660231231666578778888818998501656254556424660889999999984556535799 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MGLTQDFFPPTSELLEGGNQTSTFEFLLWGLSDQPQQQHIFFLLFLWMYVVTVAGNLLIVLAIGTDTHLHTPMYFFLASLSCADIFSTSTTVPKALVNIQTQSRSISYAGCLAQLYFFLTFGDMDIFLPATMAYDRYVAICHLLHYMMIMSLHRCAFLVTACWTLTSLLAMTRTFLIFRLSLCSAILPGFFCDLGPLMKVSCSDAQVNELVLLFLGGAVILIPFMLILVSYIRIVSAILRAPSAQGRRKAFSTCDSHLVVVALFFGTVIRAYLCPSSSSSNSVKEDTAAAVMYTVVTPLLNPFIYSMRNKDMKAAVVRLLKGRVSFSQGQ | |
| 771344123313540446130200100001014416212000010030122013312000000000120000000000010110110200210200100114334031300000110001002201000010011000000000200010002000000020020002001000000100010332000000123200300001021000101221333033133103201000000001021462321000000000000001210000000010133243443120000000131133001000020310230023003323323438 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCHHHHCCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHHSSSCCCCCCCCCCCCCSSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHCCCCCCCCC MGLTQDFFPPTSELLEGGNQTSTFEFLLWGLSDQPQQQHIFFLLFLWMYVVTVAGNLLIVLAIGTDTHLHTPMYFFLASLSCADIFSTSTTVPKALVNIQTQSRSISYAGCLAQLYFFLTFGDMDIFLPATMAYDRYVAICHLLHYMMIMSLHRCAFLVTACWTLTSLLAMTRTFLIFRLSLCSAILPGFFCDLGPLMKVSCSDAQVNELVLLFLGGAVILIPFMLILVSYIRIVSAILRAPSAQGRRKAFSTCDSHLVVVALFFGTVIRAYLCPSSSSSNSVKEDTAAAVMYTVVTPLLNPFIYSMRNKDMKAAVVRLLKGRVSFSQGQ | |||||||||||||||||||||||||
| 1 | 3emlA | 0.23 | 0.22 | 0.84 | 3.44 | Download | ----------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFT-FFCPDC--SHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL---RQ | |||||||||||||||||||
| 2 | 5tgzA | 0.23 | 0.25 | 0.85 | 2.03 | Download | ---------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFID-FHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCESVCSDIFPHI------------------DKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.21 | 0.82 | 2.05 | Download | --------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLP-------PFFCVVNTDH-------------------ILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQKYLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 4 | 4djh | 0.17 | 0.21 | 0.85 | 1.53 | Download | ---------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE--DVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALG--SAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------- | |||||||||||||||||||
| 5 | 4yay | 0.19 | 0.24 | 0.91 | 1.23 | Download | LEDKVEAQAAAEQLKTTRN-AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV------FFIITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQLDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------- | |||||||||||||||||||
| 6 | 3emlA | 0.23 | 0.22 | 0.84 | 3.54 | Download | -----------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDC--SHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.21 | 0.80 | 1.71 | Download | ----------------------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVNT-------------------DHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH----L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 8 | 2z73A | 0.19 | 0.21 | 0.89 | 2.92 | Download | ---------------ETWNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAI---GPIFGWVLCNDYISR------------DSTTRSNILCMFILG---FFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGP--LEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFD | |||||||||||||||||||
| 9 | 3emlA | 0.22 | 0.22 | 0.85 | 4.78 | Download | ----------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPML--------WNNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDC--SHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 10 | 2ydoA | 0.19 | 0.22 | 0.88 | 5.35 | Download | -------------------------------S------SVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDC--SHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEP | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
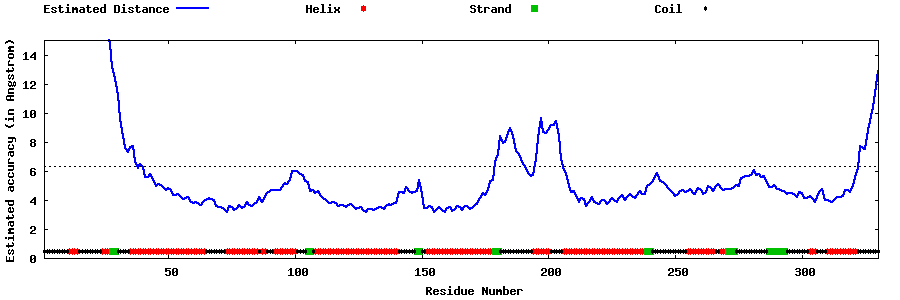 | |||
|
|
|||
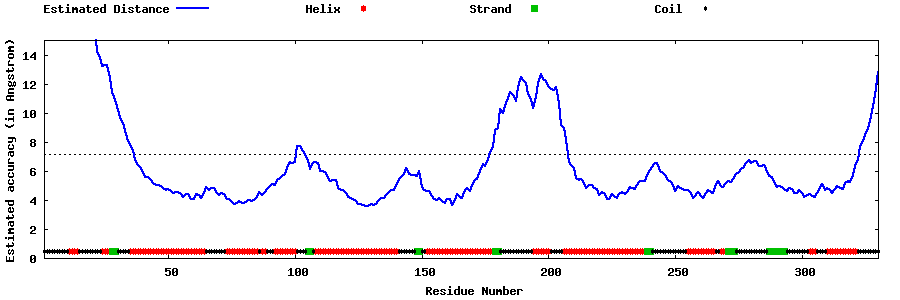 | |||
|
| |||
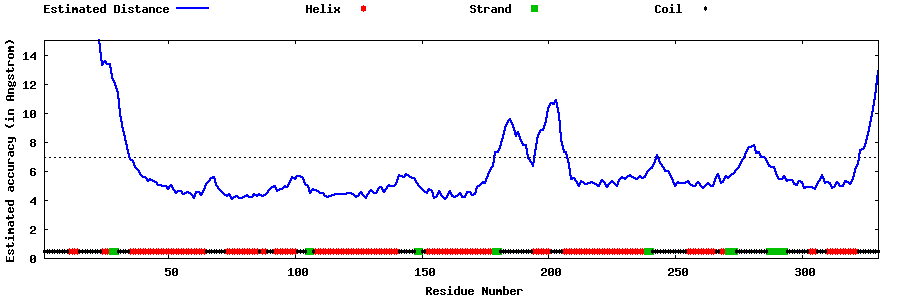 | |||
|
| |||
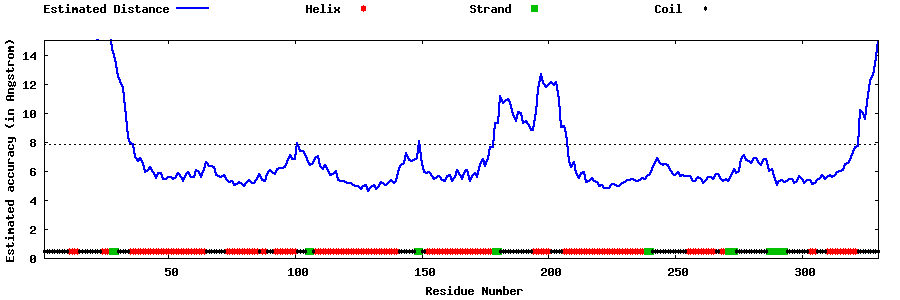 | |||
|
| |||
 | |||
|
| |||