

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MPQILIFTYLNMFYFFPPLQILAENLTMVTEFLLLGFSSLGEIQLALFVVFLFLYLVILSGNVTIISVIHLDKSLHTPMYFFLGILSTSETFYTFVILPKMLINLLSVARTISFNCCALQMFFFLGFAITNCLLLGVMGYDRYAAICHPLHYPTLMSWQVCGKLAAACAIGGFLASLTVVNLVFSLPFCSANKVNHYFCDISAVILLACTNTDVNEFVIFICGVLVLVVPFLFICVSYLCILRTILKIPSAEGRRKAFSTCASHLSVVIVHYGCASFIYLRPTANYVSNKDRLVTVTYTIVTPLLNPMVYSLRNKDVQLAIRKVLGKKGSLKLYN | |
| CCCHHHHHHHHHHHCCCHHHCCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCCC | |
| 97314567888871378422236778322576677699885269999999999999999988999687861887557489998889999888882188999998556996785899999999999999999999999986514630655558841577699999999999999999999999816078998967875258388888852576199999999999999986999999999999998228673656524326587899799987425316827899998777838998553355332354321565999999999997320230369 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MPQILIFTYLNMFYFFPPLQILAENLTMVTEFLLLGFSSLGEIQLALFVVFLFLYLVILSGNVTIISVIHLDKSLHTPMYFFLGILSTSETFYTFVILPKMLINLLSVARTISFNCCALQMFFFLGFAITNCLLLGVMGYDRYAAICHPLHYPTLMSWQVCGKLAAACAIGGFLASLTVVNLVFSLPFCSANKVNHYFCDISAVILLACTNTDVNEFVIFICGVLVLVVPFLFICVSYLCILRTILKIPSAEGRRKAFSTCASHLSVVIVHYGCASFIYLRPTANYVSNKDRLVTVTYTIVTPLLNPMVYSLRNKDVQLAIRKVLGKKGSLKLYN | |
| 44302321001002213425135403020100000000333401110013023312311332320010010033000000110200011022012003020001001653201010021011000000210000001001000000021020002003300000000000100310311011000020134130100000320002000210310021012312331331221033023200000010313622310000000010000011200100001031633553310000300232033103000010320130022003333314238 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHCCCHHHCCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCCHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCCC MPQILIFTYLNMFYFFPPLQILAENLTMVTEFLLLGFSSLGEIQLALFVVFLFLYLVILSGNVTIISVIHLDKSLHTPMYFFLGILSTSETFYTFVILPKMLINLLSVARTISFNCCALQMFFFLGFAITNCLLLGVMGYDRYAAICHPLHYPTLMSWQVCGKLAAACAIGGFLASLTVVNLVFSLPFCSANKVNHYFCDISAVILLACTNTDVNEFVIFICGVLVLVVPFLFICVSYLCILRTILKIPSAEGRRKAFSTCASHLSVVIVHYGCASFIYLRPTANYVSNKDRLVTVTYTIVTPLLNPMVYSLRNKDVQLAIRKVLGKKGSLKLYN | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.19 | 0.83 | 3.28 | Download | ----------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINC-FTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.22 | 0.84 | 2.03 | Download | ----------------------GENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL--------GWNCEKL---------QSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM----------- | |||||||||||||||||||
| 3 | 5tgzA | 0.19 | 0.22 | 0.81 | 2.09 | Download | ------------------------------GGRGENFMDIPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA-PDQARMDIELAKTLVLILVVLIICWGPLLAIMKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.19 | 0.84 | 1.51 | Download | ---------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP-------- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.23 | 0.90 | 1.25 | Download | TPPLPEMKFRHGAQAAAEQLKTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.19 | 0.83 | 3.53 | Download | -----------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.21 | 0.79 | 1.71 | Download | ----------------------------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVN----------T-------DHIL---YTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 8 | 4buoA | 0.19 | 0.19 | 0.84 | 2.98 | Download | ------------------------NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGPI----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDE----QLVYVSAAINPILYNLVSANFRQVFLSTL---------- | |||||||||||||||||||
| 9 | 3emlA | 0.17 | 0.19 | 0.83 | 4.56 | Download | ----------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.17 | 0.87 | 5.21 | Download | -------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEP | |||||||||||||||||||
| ||||||||||||||||||||||||||
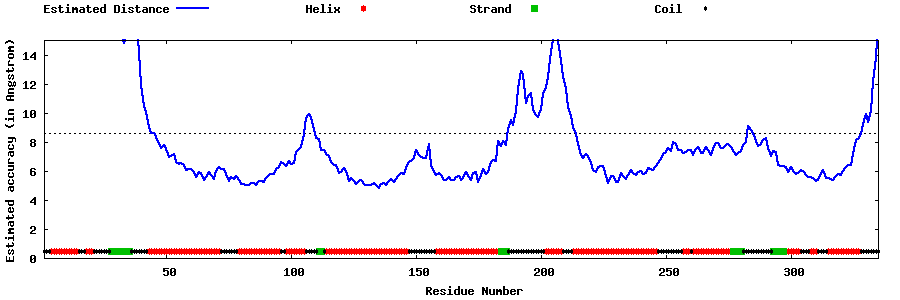
| Generated 3D models | Estimated local accuracy of models | ||
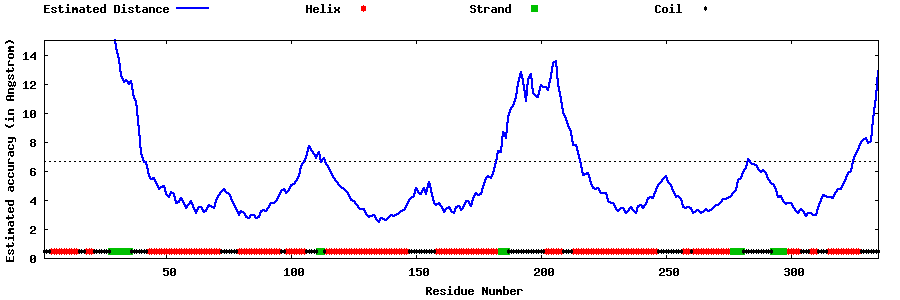 | |||
|
|
|||
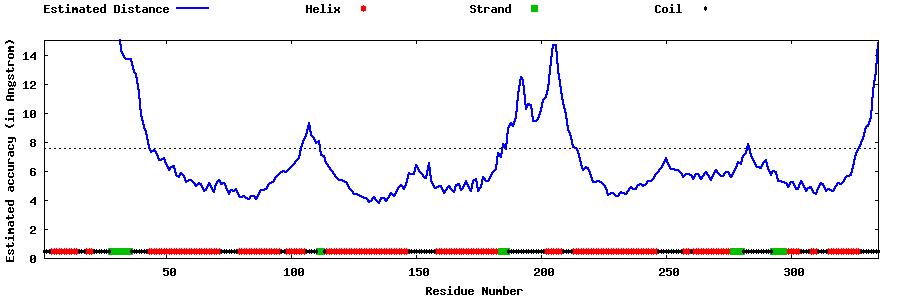 | |||
|
| |||
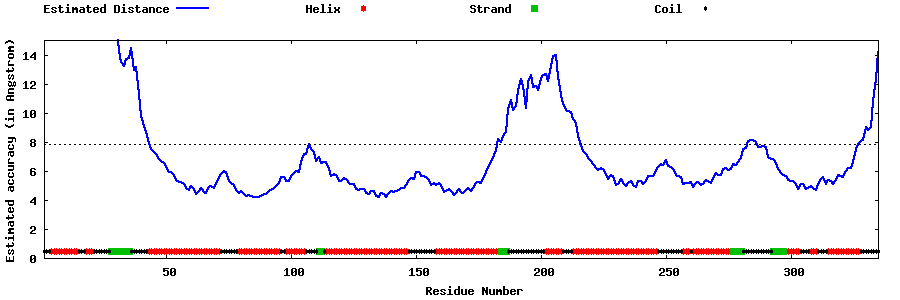 | |||
|
| |||
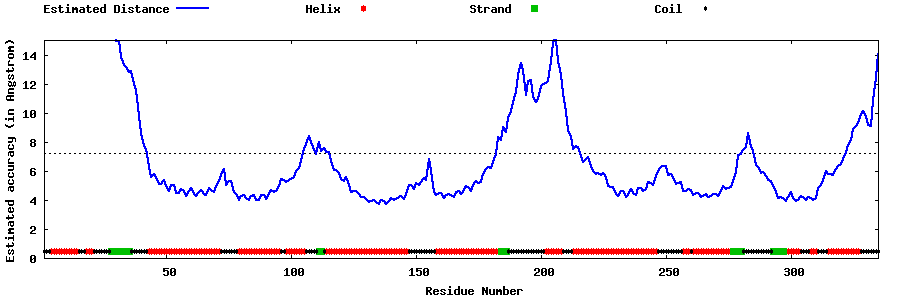 | |||
|
| |||
 | |||
|
| |||