

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MKQYSVGNQHSNYRSLLFPFLCSQMTQLTASGNQTMVTEFLFSMFPHAHRGGLLFFIPLLLIYGFILTGNLIMFIVIQVGMALHTPLYFFISVLSFLEICYTTTTIPKMLSCLISEQKSISVAGCLLQMYFFHSLGITESCVLTAMAIDRYIAICNPLRYPTIMIPKLCIQLTVGSCFCGFLLVLPEIAWISTLPFCGSNQIHQIFCDFTPVLSLACTDTFLVVIVDAIHAAEIVASFLVIALSYIRIIIVILGMHSAEGHHKAFSTCAAHLAVFLLFFGSVAVMYLRFSATYSVFWDTAIAVTFVILAPFFNPIIYSLKNKDMKEAIGRLFHYQKRAGWAGK | |
| CCCCCCCCCCCCCCSSSSHHHHHHHHHHHHCCCCSSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCSSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHHCCCCCCC | |
| 9752336778775303310100157665006887333476686699884279999999999999999988999788871887567489998889999777772288999898646996885899999999999999999999999987517761744358823677799999999999999999999999806168998968886668088787715685399999999999999999999999999999970387626576232065878998999974141378378999988788199985433664303542114659999999999985413776789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MKQYSVGNQHSNYRSLLFPFLCSQMTQLTASGNQTMVTEFLFSMFPHAHRGGLLFFIPLLLIYGFILTGNLIMFIVIQVGMALHTPLYFFISVLSFLEICYTTTTIPKMLSCLISEQKSISVAGCLLQMYFFHSLGITESCVLTAMAIDRYIAICNPLRYPTIMIPKLCIQLTVGSCFCGFLLVLPEIAWISTLPFCGSNQIHQIFCDFTPVLSLACTDTFLVVIVDAIHAAEIVASFLVIALSYIRIIIVILGMHSAEGHHKAFSTCAAHLAVFLLFFGSVAVMYLRFSATYSVFWDTAIAVTFVILAPFFNPIIYSLKNKDMKEAIGRLFHYQKRAGWAGK | |
| 6453426433430320201011242343146402020000000000333501010013023312313332320010010033001000000000001011010011030001012643201010010011000000210000000001000000020010002003300000001001100310300010002020035140100000320002000210210011223112333311310220232000000103136323100000000100000112000000010316313434100003002320330030000204201400330033234452668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCSSSSHHHHHHHHHHHHCCCCSSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCSSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHHCCCCCCC MKQYSVGNQHSNYRSLLFPFLCSQMTQLTASGNQTMVTEFLFSMFPHAHRGGLLFFIPLLLIYGFILTGNLIMFIVIQVGMALHTPLYFFISVLSFLEICYTTTTIPKMLSCLISEQKSISVAGCLLQMYFFHSLGITESCVLTAMAIDRYIAICNPLRYPTIMIPKLCIQLTVGSCFCGFLLVLPEIAWISTLPFCGSNQIHQIFCDFTPVLSLACTDTFLVVIVDAIHAAEIVASFLVIALSYIRIIIVILGMHSAEGHHKAFSTCAAHLAVFLLFFGSVAVMYLRFSATYSVFWDTAIAVTFVILAPFFNPIIYSLKNKDMKEAIGRLFHYQKRAGWAGK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.18 | 0.81 | 3.25 | Download | ------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINC-FTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ---- | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.20 | 0.82 | 2.18 | Download | -----------------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL--------GWNCEKL---------QSVCSDIFIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------------ | |||||||||||||||||||
| 3 | 4iaqA | 0.17 | 0.19 | 0.77 | 2.14 | Download | ----------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH-----------------ILYTVYSTVGAF--YFPTLLLIALYGRIYVEARSRIIQLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-------- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.17 | 0.81 | 1.52 | Download | -----------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPD----DDYWWDLFMKICVFFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------- | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.24 | 0.90 | 1.23 | Download | MRPPKLSPDSPEMRHGFVKEAQAAAEQLKTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVF-F------IITVCAFHYE--------TLPIGLGLTNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGADIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----------- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.18 | 0.81 | 3.49 | Download | -------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ---- | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.20 | 0.77 | 1.71 | Download | ------------------------------------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVN----------T-------D---HILYVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPI--HLAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-------- | |||||||||||||||||||
| 8 | 4buoA | 0.17 | 0.17 | 0.79 | 2.59 | Download | --------------------------------NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGIVDLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYI---------------------SLVYVSAAINPILYNLVSANFRQVFLSTL----------- | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.18 | 0.81 | 4.69 | Download | ------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDV----------VPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ---- | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.18 | 0.85 | 5.25 | Download | ---------------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFCVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPF | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
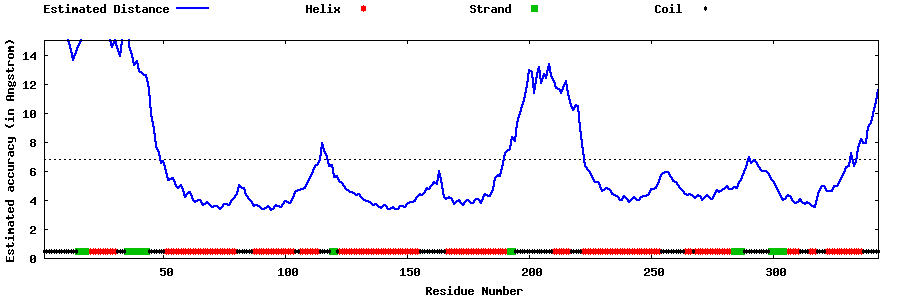 | |||
|
| |||
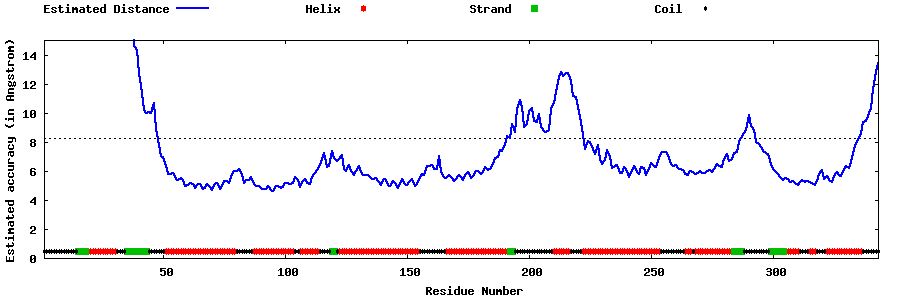 | |||
|
| |||
 | |||
|
| |||