

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MSGENVTRVGTFILVGFPTAPGLQYLLFLLFLLTYLFVLVENLAIILTVWSSTSLHRPMYYFLSSMSFLEIWYVSDITPKMLEGFLLQQKRISFVGCMTQLYFFSSLVCTECVLLASMAYDRYVAICHPLRYHVLVTPGLCLQLVGFSFVSGFTISMIKVCFISSVTFCGSNVLNHFFCDISPILKLACTDFSTAELVDFILAFIILVFPLLATMLSYAHITLAVLRIPSATGCWRAFFTCASHLTVVTVFYTALLFMYVRPQAIDSRSSNKLISVLYTVITPILNPLIYCLRNKEFKNALKKAFGLTSCAVEGRLSSLLELHLQIHSQPL | |
| CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHCCCCCC | |
| 9998883235667856898843599999999999999999899998988618886675899987899986633505889998987059978868999999999999999999999999875046407553488314787999999999999999999999998061588999578852580888877157862999999999999999989999999999999980386636665321065878998999973151368178999888788589986114142123653215659999999999860426533431146642320257889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MSGENVTRVGTFILVGFPTAPGLQYLLFLLFLLTYLFVLVENLAIILTVWSSTSLHRPMYYFLSSMSFLEIWYVSDITPKMLEGFLLQQKRISFVGCMTQLYFFSSLVCTECVLLASMAYDRYVAICHPLRYHVLVTPGLCLQLVGFSFVSGFTISMIKVCFISSVTFCGSNVLNHFFCDISPILKLACTDFSTAELVDFILAFIILVFPLLATMLSYAHITLAVLRIPSATGCWRAFFTCASHLTVVTVFYTALLFMYVRPQAIDSRSSNKLISVLYTVITPILNPLIYCLRNKEFKNALKKAFGLTSCAVEGRLSSLLELHLQIHSQPL | |
| 8665130300100000004324010100230333133133333100100100330000000101000010110100200200010015432000300000110000000000100000000000000100200030033000000000001003003010100020301341201000003200030002203110111021013113212300330331001000103146223100000000200010132001000010325323434100000103300330030000103201400330043340124430330132304344435 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHCCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHCCCCCC MSGENVTRVGTFILVGFPTAPGLQYLLFLLFLLTYLFVLVENLAIILTVWSSTSLHRPMYYFLSSMSFLEIWYVSDITPKMLEGFLLQQKRISFVGCMTQLYFFSSLVCTECVLLASMAYDRYVAICHPLRYHVLVTPGLCLQLVGFSFVSGFTISMIKVCFISSVTFCGSNVLNHFFCDISPILKLACTDFSTAELVDFILAFIILVFPLLATMLSYAHITLAVLRIPSATGCWRAFFTCASHLTVVTVFYTALLFMYVRPQAIDSRSSNKLISVLYTVITPILNPLIYCLRNKEFKNALKKAFGLTSCAVEGRLSSLLELHLQIHSQPL | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.19 | 0.84 | 3.37 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------------------- | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.21 | 0.85 | 2.05 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL--------GWNCEKL---------QSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM--------------------------- | |||||||||||||||||||
| 3 | 5tgzA | 0.19 | 0.21 | 0.82 | 2.00 | Download | ----------GGRGENFMDIPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQADIELAKTLVLILVVLIICWGPLLAIMVDVFGKMNKLIKTVFAFCLCLLNSTVNPIIYALRSKDLRHAFRSMF-------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.22 | 0.85 | 1.53 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------ | |||||||||||||||||||
| 5 | 4yay | 0.14 | 0.21 | 0.85 | 1.24 | Download | KTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV-FF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------------------------- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.19 | 0.84 | 3.54 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------------------- | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.22 | 0.79 | 1.71 | Download | --------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWFIISLVMPIH--LAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.17 | 0.20 | 0.82 | 2.64 | Download | ----NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPI------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQRAVVIAFVVCWLPYHVRRLMFCYI-----------SDEQ-------WTYVSAAINPILYNLVSANFRQVFLSTL-------------------------- | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.19 | 0.84 | 4.74 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ------------------- | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.19 | 0.91 | 5.88 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFKAAAAENLYF----- | |||||||||||||||||||
| ||||||||||||||||||||||||||
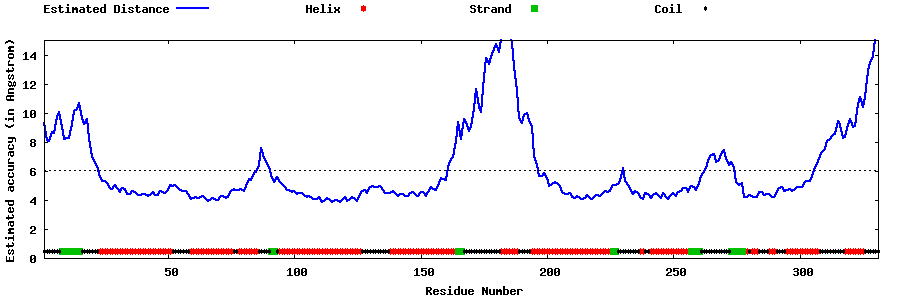
| Generated 3D models | Estimated local accuracy of models | ||
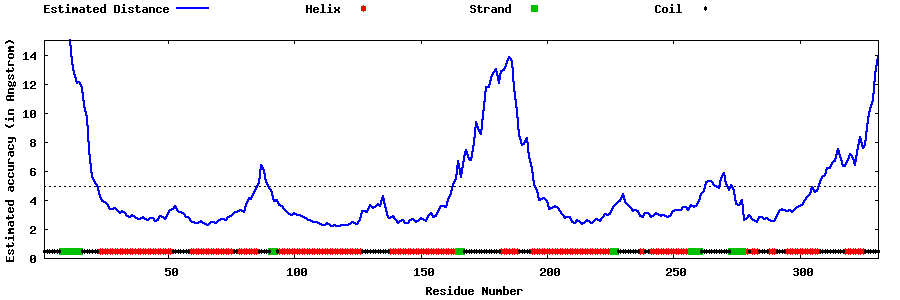 | |||
|
|
|||
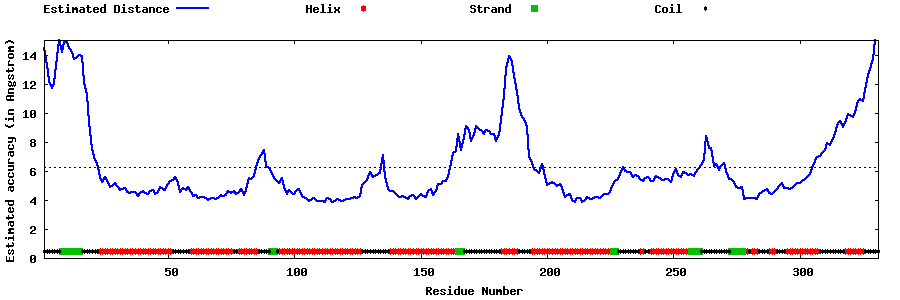 | |||
|
| |||
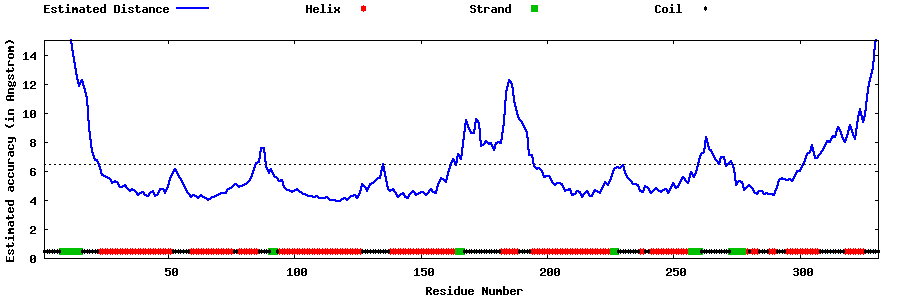 | |||
|
| |||
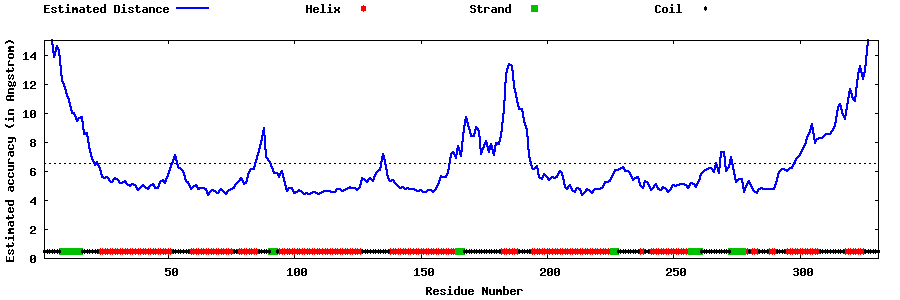 | |||
|
| |||
 | |||
|
| |||