

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MNITLYSFLYVLRLNEGNSTGRNIDERKKMQGENFTIWSIFFLEGFSQYPGLEVVLFVFSLVMYLTTLLGNSTLILITILDSRLKTPMYLFLGNLSFMDICYTSASVPTLLVNLLSSQKTIIFSGCAVQMYLSLAMGSTECVLLAVMAYDRYVAICNPLRYSIIMNRCVCARMATVSWVTGCLTALLETSFALQIPLCGNLIDHFTCEILAVLKLACTSSLLMNTIMLVVSILLLPIPMLLVCISYIFILSTILRITSAEGRNKAFSTCGAHLTVVILYYGAALSMYLKPSSSNAQKIDKIISLLYGVLTPMLNPIIYSLRNKEVKDAMKKLLGKITLHQTHEHL | |
| CCCCHHHHHHHHHHCCCCHHHHCCCHHHCCCCCCCCSSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCSSCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCCCCC | |
| 970302457887641332043216332015577882236778965899851799999999999999999889997888628875674898887899987888701789999984789965738999999999999999999999999764066405554576236887999999999999999999999998360777896787624818889886147609999999999999999799999999999999831866043651432558689979998731540780789999877781898644134333124643156499999999998626324101469 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MNITLYSFLYVLRLNEGNSTGRNIDERKKMQGENFTIWSIFFLEGFSQYPGLEVVLFVFSLVMYLTTLLGNSTLILITILDSRLKTPMYLFLGNLSFMDICYTSASVPTLLVNLLSSQKTIIFSGCAVQMYLSLAMGSTECVLLAVMAYDRYVAICNPLRYSIIMNRCVCARMATVSWVTGCLTALLETSFALQIPLCGNLIDHFTCEILAVLKLACTSSLLMNTIMLVVSILLLPIPMLLVCISYIFILSTILRITSAEGRNKAFSTCGAHLTVVILYYGAALSMYLKPSSSNAQKIDKIISLLYGVLTPMLNPIIYSLRNKEVKDAMKKLLGKITLHQTHEHL | |
| 641201000301324344343541443540454020201100000003324001100130233123123323200100100230000000102000000010000210300000116522010100120012013002100200010020000000210200000033001000110010002103100000010110213000000031000000021033000112331133133122000201120020001031361232011000000000000110000000103163264230000021023203300300001031013002200242214443644 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCHHHHHHHHHHCCCCHHHHCCCHHHCCCCCCCCSSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCSSCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCCCCC MNITLYSFLYVLRLNEGNSTGRNIDERKKMQGENFTIWSIFFLEGFSQYPGLEVVLFVFSLVMYLTTLLGNSTLILITILDSRLKTPMYLFLGNLSFMDICYTSASVPTLLVNLLSSQKTIIFSGCAVQMYLSLAMGSTECVLLAVMAYDRYVAICNPLRYSIIMNRCVCARMATVSWVTGCLTALLETSFALQIPLCGNLIDHFTCEILAVLKLACTSSLLMNTIMLVVSILLLPIPMLLVCISYIFILSTILRITSAEGRNKAFSTCGAHLTVVILYYGAALSMYLKPSSSNAQKIDKIISLLYGVLTPMLNPIIYSLRNKEVKDAMKKLLGKITLHQTHEHL | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.19 | 0.80 | 3.23 | Download | -------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----- | |||||||||||||||||||
| 2 | 5tgzA | 0.23 | 0.24 | 0.81 | 2.12 | Download | ------------------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCESVCSDIFPHI------------------DKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSM------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.20 | 0.19 | 0.76 | 2.10 | Download | -----------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVN----------------TDHILYTVYSTVGAF---YFPTLLLIALYGRIYVEARSRIIQKYAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--------- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.19 | 0.81 | 1.52 | Download | ------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR--EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.22 | 0.90 | 1.24 | Download | LEDKSSPEMFRHGFDEGKVKEAQAAAEQLKTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------------ | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.19 | 0.80 | 3.53 | Download | --------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----- | |||||||||||||||||||
| 7 | 4iaq | 0.22 | 0.20 | 0.76 | 1.69 | Download | --------------------------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASE---------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH-L-AI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--------- | |||||||||||||||||||
| 8 | 4buoA | 0.18 | 0.17 | 0.82 | 2.67 | Download | ---------------------------------NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPTPI----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTNALVYVSAAINPILYNLVSANFRQVFLSTL------------ | |||||||||||||||||||
| 9 | 3emlA | 0.19 | 0.19 | 0.81 | 4.42 | Download | -------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQSGCGEGQVAC--------LFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ----- | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.18 | 0.84 | 5.17 | Download | ----------------------------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEPFK | |||||||||||||||||||
| ||||||||||||||||||||||||||
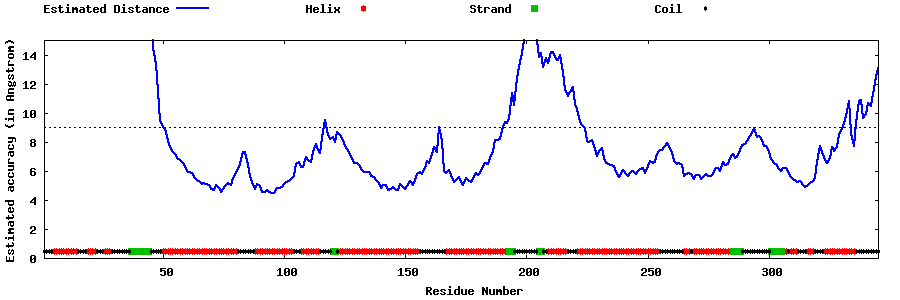
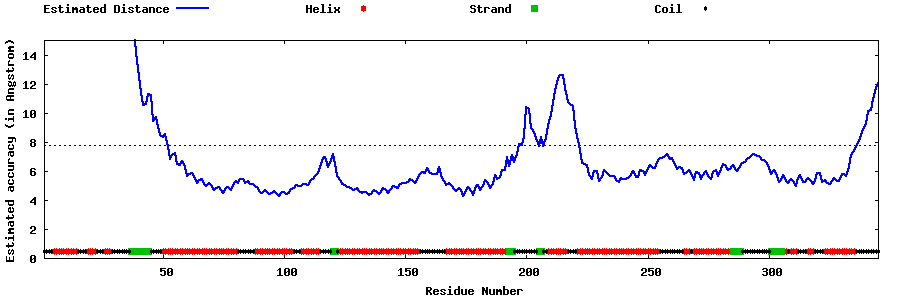
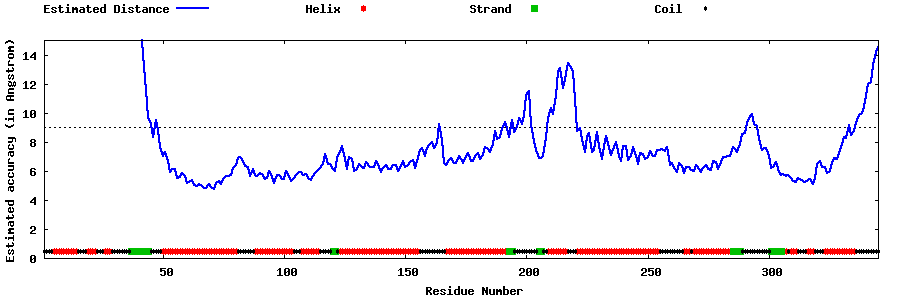
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||