

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MENQSSISEFFLRGISAPPEQQQSLFGIFLCMYLVTLTGNLLIILAIGSDLHLHTPMYFFLANLSFVDMGLTSSTVTKMLVNIQTRHHTISYTGCLTQMYFFLMFGDLDSFFLAAMAYDRYVAICHPLCYSTVMRPQVCALMLALCWVLTNIVALTHTFLMARLSFCVTGEIAHFFCDITPVLKLSCSDTHINEMMVFVLGGTVLIVPFLCIVTSYIHIVPAILRVRTRGGVGKAFSTCSSHLCVVCVFYGTLFSAYLCPPSIASEEKDIAAAAMYTIVTPMLNPFIYSLRNKDMKGALKRLFSHRSIVSS | |
| CCCCCCCHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHSSSCCCCCCCCCCCCSSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHHCCCCCC | |
| 99886346562007999810558999999999999999899999999628875552999988799875451314879887998438966858999999999999999899999999875055016654378431760899999999999999999999998751479999468926786999766164425878999999999999879999999999999841136778867620560778999999867043367078987889888089985016562545554236728899999999841405479 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MENQSSISEFFLRGISAPPEQQQSLFGIFLCMYLVTLTGNLLIILAIGSDLHLHTPMYFFLANLSFVDMGLTSSTVTKMLVNIQTRHHTISYTGCLTQMYFFLMFGDLDSFFLAAMAYDRYVAICHPLCYSTVMRPQVCALMLALCWVLTNIVALTHTFLMARLSFCVTGEIAHFFCDITPVLKLSCSDTHINEMMVFVLGGTVLIVPFLCIVTSYIHIVPAILRVRTRGGVGKAFSTCSSHLCVVCVFYGTLFSAYLCPPSIASEEKDIAAAAMYTIVTPMLNPFIYSLRNKDMKGALKRLFSHRSIVSS | |
| 87241210200001014316212000110030122023312100000100230000000000010110111200310300100114344031300000110001002201000000021000000110200000002000000120020002002100000200010372400000012320030000102101010122133313323300330101000000102146232100000000000000121002000000122314432200000001322330010000203102300230033342358 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHSSSCCCCCCCCCCCCSSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHHCCCCCC MENQSSISEFFLRGISAPPEQQQSLFGIFLCMYLVTLTGNLLIILAIGSDLHLHTPMYFFLANLSFVDMGLTSSTVTKMLVNIQTRHHTISYTGCLTQMYFFLMFGDLDSFFLAAMAYDRYVAICHPLCYSTVMRPQVCALMLALCWVLTNIVALTHTFLMARLSFCVTGEIAHFFCDITPVLKLSCSDTHINEMMVFVLGGTVLIVPFLCIVTSYIHIVPAILRVRTRGGVGKAFSTCSSHLCVVCVFYGTLFSAYLCPPSIASEEKDIAAAAMYTIVTPMLNPFIYSLRNKDMKGALKRLFSHRSIVSS | |||||||||||||||||||||||||
| 1 | 3emlA | 0.21 | 0.22 | 0.89 | 3.74 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.26 | 0.90 | 2.14 | Download | -ENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFID-FHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-----------------GWNCEKLQSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 3 | 4iaqA | 0.19 | 0.23 | 0.83 | 2.06 | Download | ----------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISILYTVYSTVGAF---------------------------------------YFPTLLLIALYGRIYVEARSRIIQKYAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.24 | 0.90 | 1.54 | Download | -----------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------ | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.22 | 0.91 | 1.23 | Download | TTRNAYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHR-NVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------- | |||||||||||||||||||
| 6 | 3emlA | 0.22 | 0.22 | 0.89 | 3.91 | Download | -------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 7 | 4iaq | 0.22 | 0.25 | 0.85 | 1.71 | Download | -----------------SLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVNT-----------------DHI---LYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH---LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 8 | 2z73A | 0.19 | 0.24 | 0.96 | 3.01 | Download | WYNPSIVVHPHWREFDQVPDAVYYSLGIFIGCGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFG------WGAYTLEGVLCD-------YISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQ | |||||||||||||||||||
| 9 | 3emlA | 0.22 | 0.22 | 0.89 | 5.35 | Download | -----------IM-------GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 10 | 2ydoA | 0.19 | 0.22 | 0.93 | 6.00 | Download | ---------------S------SVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQ | |||||||||||||||||||
| ||||||||||||||||||||||||||
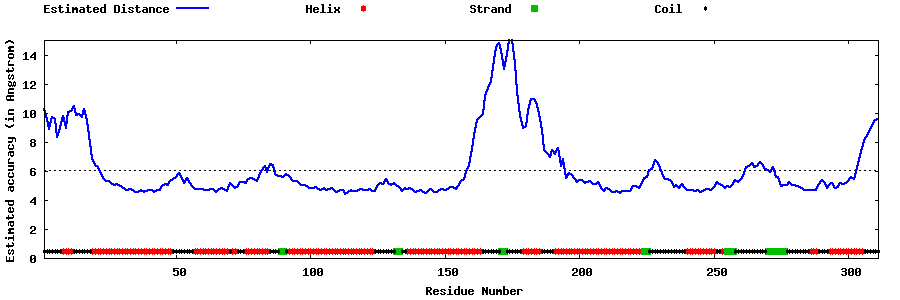
| Generated 3D models | Estimated local accuracy of models | ||
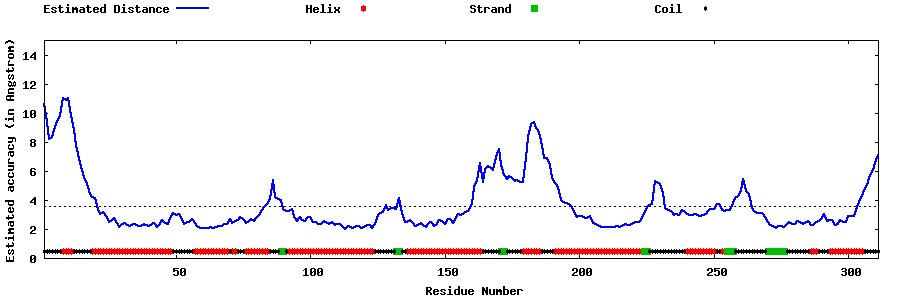 | |||
|
|
|||
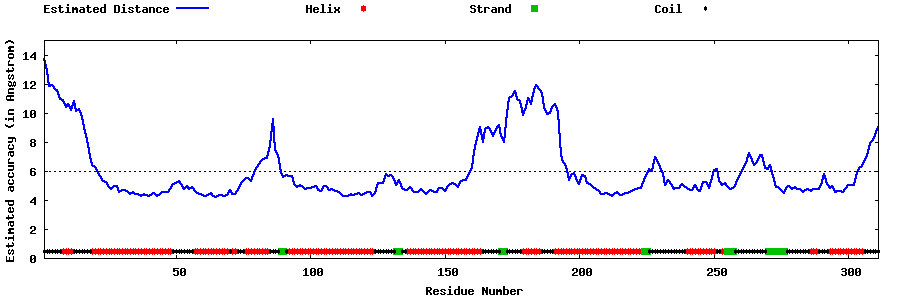 | |||
|
| |||
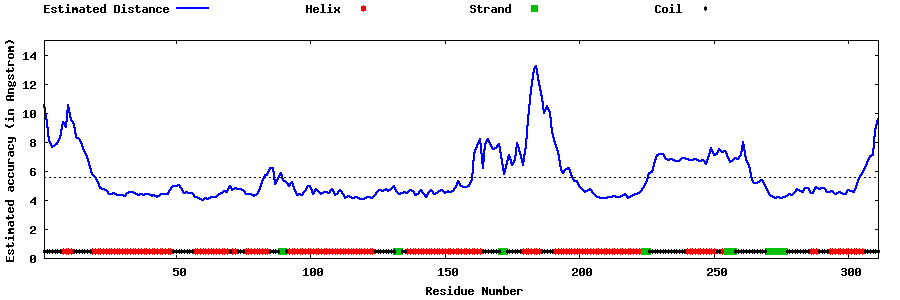 | |||
|
| |||
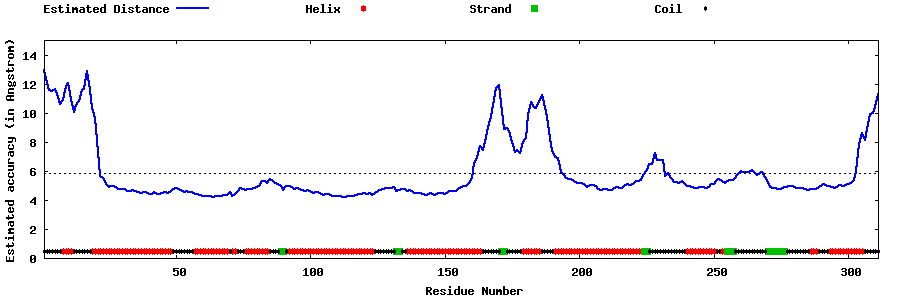 | |||
|
| |||
 | |||
|
| |||