

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MEGFYLRRSHELQGMGKPGRVNQTTVSDFLLLGLSEWPEEQPLLFGIFLGMYLVTMVGNLLIILAISSDPHLHTPMYFFLANLSLTDACFTSASIPKMLANIHTQSQIISYSGCLAQLYFLLMFGGLDNCLLAVMAYDRYVAICQPLHYSTSMSPQLCALMLGVCWVLTNCPALMHTLLLTRVAFCAQKAIPHFYCDPSALLKLACSDTHVNELMIITMGLLFLTVPLLLIVFSYVRIFWAVFVISSPGGRWKAFSTCGSHLTVVLLFYGSLMGVYLLPPSTYSTERESRAAVLYMVIIPTLNPFIYSLRNRDMKEALGKLFVSGKTFFL | |
| CCCHHHCCHHHHHHHHCCCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCCHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHHCCCCCC | |
| 963020210655554016676786265451116899831668999999999999999899999999717875563999988799875451325879888988358967738899999999999999999999999875132336754378431760899999999999999999999999732689999358826786999866164427989999999999999889999999999998831235778867620671768998999867043267178977889988189985006462545653146618899999999963523469 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MEGFYLRRSHELQGMGKPGRVNQTTVSDFLLLGLSEWPEEQPLLFGIFLGMYLVTMVGNLLIILAISSDPHLHTPMYFFLANLSLTDACFTSASIPKMLANIHTQSQIISYSGCLAQLYFLLMFGGLDNCLLAVMAYDRYVAICQPLHYSTSMSPQLCALMLGVCWVLTNCPALMHTLLLTRVAFCAQKAIPHFYCDPSALLKLACSDTHVNELMIITMGLLFLTVPLLLIVFSYVRIFWAVFVISSPGGRWKAFSTCGSHLTVVLLFYGSLMGVYLLPPSTYSTERESRAAVLYMVIIPTLNPFIYSLRNRDMKEALGKLFVSGKTFFL | |
| 533022332331312340446130200100001014416212000010030122013312000000100120000000000010110110200310300100114334030200000110000002101000000011000000010200000002000000020020001001000000000010362400000012320030000102101010122133303323300220101000000102146232100000000000000111001000000021214332200000001211330010000203102300230034342014 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHCCHHHHHHHHCCCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCCHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHHCCCCCC MEGFYLRRSHELQGMGKPGRVNQTTVSDFLLLGLSEWPEEQPLLFGIFLGMYLVTMVGNLLIILAISSDPHLHTPMYFFLANLSLTDACFTSASIPKMLANIHTQSQIISYSGCLAQLYFLLMFGGLDNCLLAVMAYDRYVAICQPLHYSTSMSPQLCALMLGVCWVLTNCPALMHTLLLTRVAFCAQKAIPHFYCDPSALLKLACSDTHVNELMIITMGLLFLTVPLLLIVFSYVRIFWAVFVISSPGGRWKAFSTCGSHLTVVLLFYGSLMGVYLLPPSTYSTERESRAAVLYMVIIPTLNPFIYSLRNRDMKEALGKLFVSGKTFFL | |||||||||||||||||||||||||
| 1 | 3emlA | 0.22 | 0.21 | 0.84 | 3.39 | Download | -------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH---VL | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.21 | 0.85 | 2.03 | Download | ------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFID-FHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-----------------GWNCEKLQSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM--------- | |||||||||||||||||||
| 3 | 4iaqA | 0.21 | 0.20 | 0.78 | 2.00 | Download | -----------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISILYTVYSTVGAF---------------------------------------YFPTLLLIALYGRIYVEARSRIIQKYLLMERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.21 | 0.85 | 1.53 | Download | ------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------ | |||||||||||||||||||
| 5 | 4yay | 0.18 | 0.24 | 0.91 | 1.23 | Download | LEDLVGQIEAQAAAEQLKTTRN-AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRN-VFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------- | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.21 | 0.84 | 3.66 | Download | --------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 7 | 4iaq | 0.23 | 0.22 | 0.79 | 1.71 | Download | -------------------------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWLPFFLVMPIH---LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 8 | 2z73A | 0.20 | 0.22 | 0.89 | 2.70 | Download | ----------------ETWWYNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFG------WGAYTLEGVLCD-------YISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTF-------P | |||||||||||||||||||
| 9 | 3emlA | 0.21 | 0.21 | 0.84 | 4.81 | Download | -------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.21 | 0.87 | 5.36 | Download | ----------------------------------S------SVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHNLYF- | |||||||||||||||||||
| ||||||||||||||||||||||||||
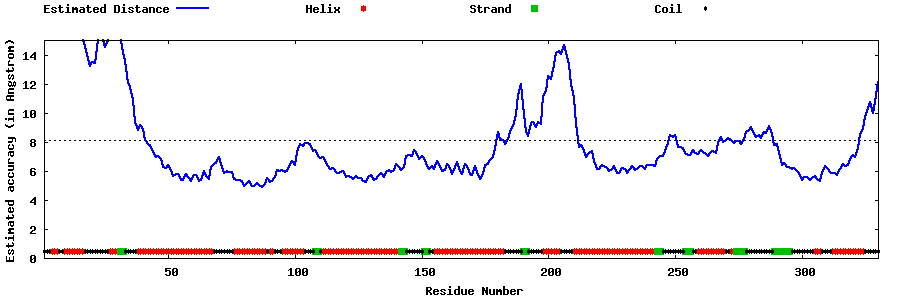
| Generated 3D models | Estimated local accuracy of models | ||
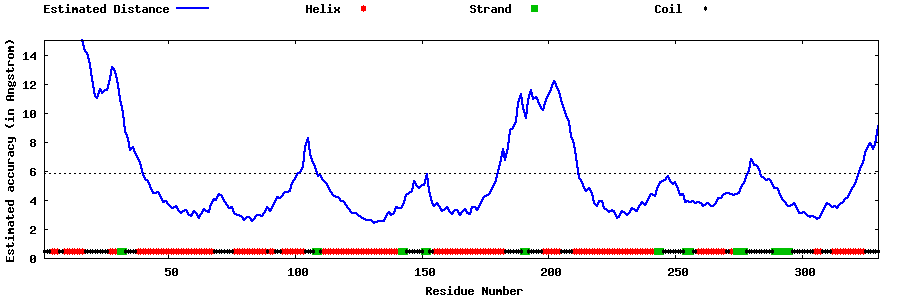 | |||
|
|
|||
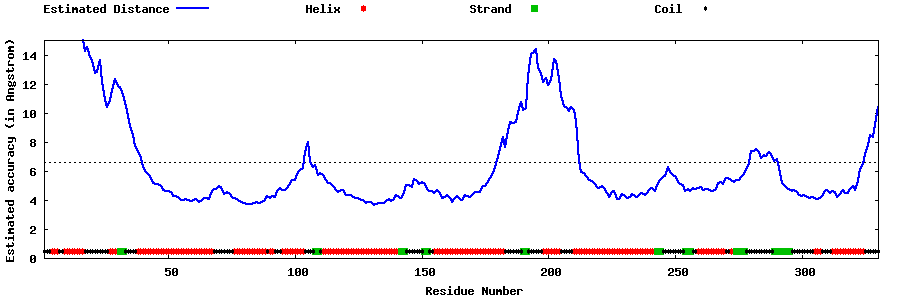 | |||
|
| |||
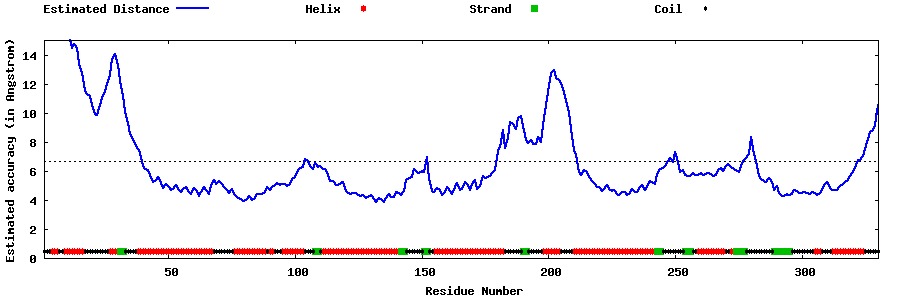 | |||
|
| |||
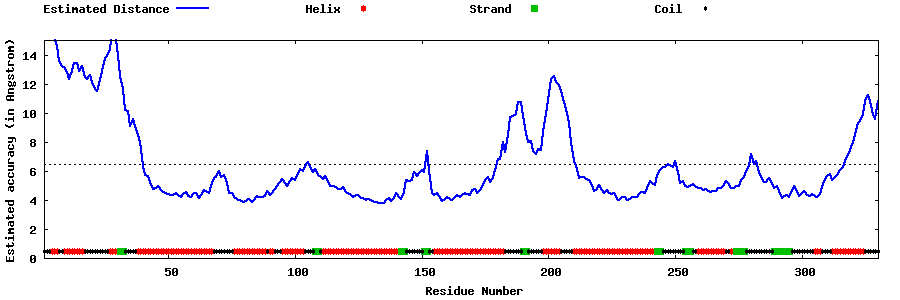 | |||
|
| |||
 | |||
|
| |||