

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MSYFYRLKLMKEAVLVKLPFTSLPLLLQTLSRKSRDMEIKNYSSSTSGFILLGLSSNPQLQKPLFAIFLIMYLLAAVGNVLIIPAIYSDPRLHTPMYFFLSNLSFMDICFTTVIVPKMLVNFLSETKVISYVGCLAQMYFFMAFGNTDSYLLASMAIDRLVAICNPLHYDVVMKPRHCLLMLLGSCSISHLHSLFRVLLMSRLSFCASHIIKHFFCDTQPVLKLSCSDTSSSQMVVMTETLAVIVTPFLCIIFSYLRIMVTVLRIPSAAGKWKAFSTCGSHLTAVALFYGSIIYVYFRPLSMYSVVRDRVATVMYTVVTPMLNPFIYSLRNKDMKRGLKKLQDRIYR | |
| CCCHHHHHHHCCCSSSSCCCCCHHHHHHHHHHHHHHHCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCHHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHCC | |
| 94036646412241664242211588874233441314767871026760317998830668999999999999999899999999638865563999987789874360326879887998438966868999999999999999999999999875166406543177402772899999999999999999999998632489999008926786898765165435878999999999999989999999999999850045778866322561878999999857031478278987889888189986116562445664236628899999999986529 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MSYFYRLKLMKEAVLVKLPFTSLPLLLQTLSRKSRDMEIKNYSSSTSGFILLGLSSNPQLQKPLFAIFLIMYLLAAVGNVLIIPAIYSDPRLHTPMYFFLSNLSFMDICFTTVIVPKMLVNFLSETKVISYVGCLAQMYFFMAFGNTDSYLLASMAIDRLVAICNPLHYDVVMKPRHCLLMLLGSCSISHLHSLFRVLLMSRLSFCASHIIKHFFCDTQPVLKLSCSDTSSSQMVVMTETLAVIVTPFLCIIFSYLRIMVTVLRIPSAAGKWKAFSTCGSHLTAVALFYGSIIYVYFRPLSMYSVVRDRVATVMYTVVTPMLNPFIYSLRNKDMKRGLKKLQDRIYR | |
| 44203303224402114021111210021023123404443341300100000014426212000000030122023312100000000120000000000010010110200210200100114334031300000110010002100000000011000000000200010002000000120020002001000000100010372401000012320020000102100010123133313312300130101000000102135132100000000000000111001000000011234432200000000321330020000203101300230032028 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHCCCSSSSCCCCCHHHHHHHHHHHHHHHCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCHHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSSCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHCC MSYFYRLKLMKEAVLVKLPFTSLPLLLQTLSRKSRDMEIKNYSSSTSGFILLGLSSNPQLQKPLFAIFLIMYLLAAVGNVLIIPAIYSDPRLHTPMYFFLSNLSFMDICFTTVIVPKMLVNFLSETKVISYVGCLAQMYFFMAFGNTDSYLLASMAIDRLVAICNPLHYDVVMKPRHCLLMLLGSCSISHLHSLFRVLLMSRLSFCASHIIKHFFCDTQPVLKLSCSDTSSSQMVVMTETLAVIVTPFLCIIFSYLRIMVTVLRIPSAAGKWKAFSTCGSHLTAVALFYGSIIYVYFRPLSMYSVVRDRVATVMYTVVTPMLNPFIYSLRNKDMKRGLKKLQDRIYR | |||||||||||||||||||||||||
| 1 | 3emlA | 0.21 | 0.19 | 0.80 | 3.21 | Download | ---------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVR | |||||||||||||||||||
| 2 | 5tgzA | 0.18 | 0.18 | 0.81 | 1.96 | Download | --------------------------------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-----------------GWNCEKLQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------ | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.18 | 0.78 | 1.92 | Download | -------------------------------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL-----------------PPFFWRQA----SECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQKYLLMERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.21 | 0.81 | 1.53 | Download | --------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.23 | 0.92 | 1.23 | Download | LTKMRAAALDAQKATPPSPEMRFDILVGEAQAAAEQLKTTRN-AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV-FF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----- | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.19 | 0.79 | 3.42 | Download | ----------------------------------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.19 | 0.75 | 1.70 | Download | -----------------------------------------------------------WKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIAAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAIF-DFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF--- | |||||||||||||||||||
| 8 | 4ea3A | 0.16 | 0.16 | 0.75 | 2.81 | Download | --------------------------------------------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAIMGSAQVEDEIECLVEIPTPQDYWGPVFAICIFLFSF-----------------IVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLG-----VQPSSETAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR-------- | |||||||||||||||||||
| 9 | 3emlA | 0.21 | 0.19 | 0.80 | 4.46 | Download | ---------------------------------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 10 | 3sn6R | 0.17 | 0.21 | 0.98 | 4.58 | Download | KPVYDSLDAVRRAALINMVFQMGETGVAGFTNSLRMLQQKNRAKRVITTFRTGTWDAEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRQEAINCYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGRCLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRK-EVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC---- | |||||||||||||||||||
| ||||||||||||||||||||||||||
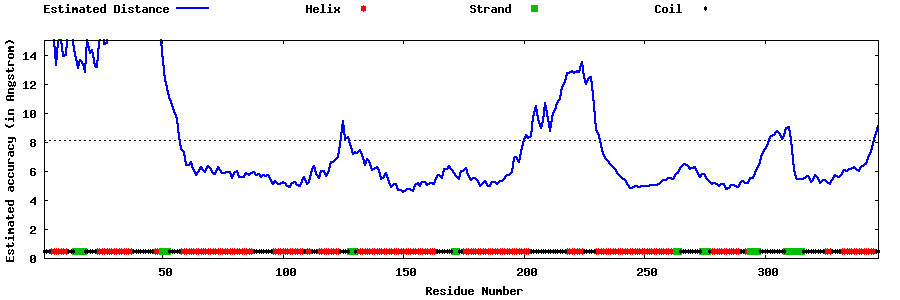
| Generated 3D models | Estimated local accuracy of models | ||
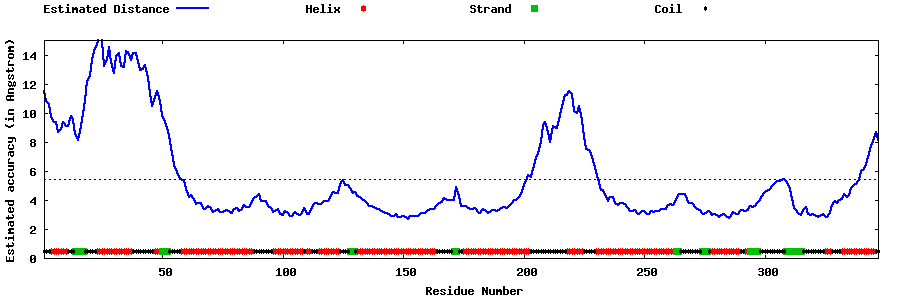 | |||
|
|
|||
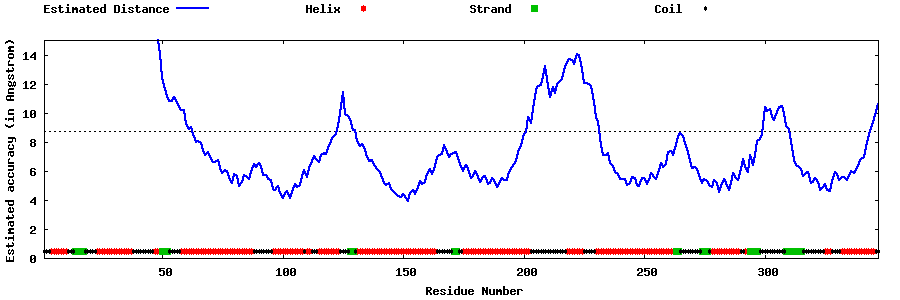 | |||
|
| |||
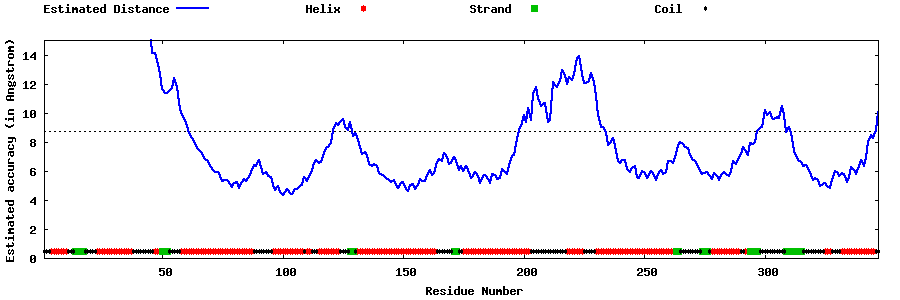 | |||
|
| |||
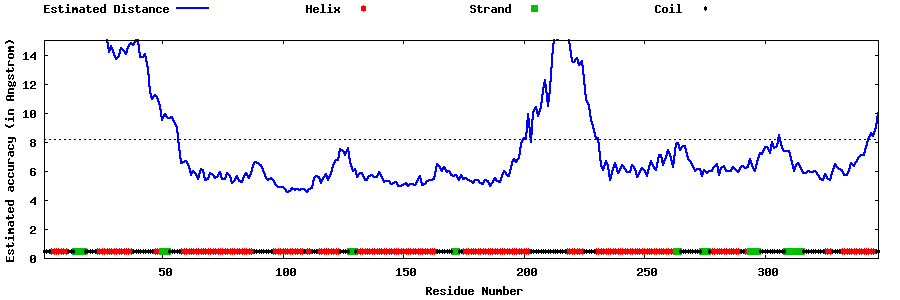 | |||
|
| |||
 | |||
|
| |||