

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MSIINTSYVEITTFFLVGMPGLEYAHIWISIPICSMYLIAILGNGTILFIIKTEPSLHEPMYYFLSMLAMSDLGLSLSSLPTVLSIFLFNAPEISSNACFAQEFFIHGFSVLESSVLLIMSFDRFLAIHNPLRYTSILTTVRVAQIGIVFSFKSMLLVLPFPFTLRNLRYCKKNQLSHSYCLHQDVMKLACSDNRIDVIYGFFGALCLMVDFILIAVSYTLILKTVLGIASKKEQLKALNTCVSHICAVIIFYLPIINLAVVHRFARHVSPLINVLMANVLLLVPPLTNPIVYCVKTKQIRVRVVAKLCQRKI | |
| CCCCCCCCCCCCHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCCSSSCCHHHHHHHHHHHHCCCCC | |
| 9998999880400116799897668899999999999999999999999996678753208999999999999999967999999995699878888999999999999999999999998353897466545760188899999999999999999999999995199999895787322006578871479537469999999999998999999999999998078898788998760699999999999999999733136898995899999999974774536800006579999999998602779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MSIINTSYVEITTFFLVGMPGLEYAHIWISIPICSMYLIAILGNGTILFIIKTEPSLHEPMYYFLSMLAMSDLGLSLSSLPTVLSIFLFNAPEISSNACFAQEFFIHGFSVLESSVLLIMSFDRFLAIHNPLRYTSILTTVRVAQIGIVFSFKSMLLVLPFPFTLRNLRYCKKNQLSHSYCLHQDVMKLACSDNRIDVIYGFFGALCLMVDFILIAVSYTLILKTVLGIASKKEQLKALNTCVSHICAVIIFYLPIINLAVVHRFARHVSPLINVLMANVLLLVPPLTNPIVYCVKTKQIRVRVVAKLCQRKI | |
| 4443242423132020000012132020001102202220332121000002014100200020000002002000300202000000021350314000000120131033012001100200000002101000000342001000000221201120210102103104620000000102100100023030011101201313332231032011100200010014601320120010000000003211321010010143101100011032100300330030000203300410031025448 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCCSSSCCHHHHHHHHHHHHCCCCC MSIINTSYVEITTFFLVGMPGLEYAHIWISIPICSMYLIAILGNGTILFIIKTEPSLHEPMYYFLSMLAMSDLGLSLSSLPTVLSIFLFNAPEISSNACFAQEFFIHGFSVLESSVLLIMSFDRFLAIHNPLRYTSILTTVRVAQIGIVFSFKSMLLVLPFPFTLRNLRYCKKNQLSHSYCLHQDVMKLACSDNRIDVIYGFFGALCLMVDFILIAVSYTLILKTVLGIASKKEQLKALNTCVSHICAVIIFYLPIINLAVVHRFARHVSPLINVLMANVLLLVPPLTNPIVYCVKTKQIRVRVVAKLCQRKI | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.22 | 0.88 | 2.46 | Download | ---------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDV----------VPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.23 | 0.86 | 2.07 | Download | -----------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVV--AFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIF-------------PHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 3 | 5tgzA | 0.20 | 0.23 | 0.89 | 1.97 | Download | -----GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID-----------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVF-GKMNIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 4 | 3d4s | 0.19 | 0.19 | 0.88 | 1.55 | Download | ------------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRA--TH--QEAINCYAEET----CCDFFTNQAYAIASSIVSYVPLVIMVFVYSRVFQEAKRQLNILKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL--- | |||||||||||||||||||
| 5 | 3uonA | 0.20 | 0.20 | 0.87 | 1.13 | Download | ----------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFI--VGVRTVEDGECYIQFF---------SNAAVTFGTIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.22 | 0.88 | 2.65 | Download | -----------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 3d4s | 0.19 | 0.19 | 0.88 | 1.69 | Download | -------------------------VVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMWYRAT---HQ--EAINCYAEE----TCCDFFTNQAYAISSIVSFYVPLVIMVFVYSRVFQEAKRQLNILKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNIRKEV-YILLNWIGYVNSGFNPLIYCR-SPDFRIAFQELLCL--- | |||||||||||||||||||
| 8 | 4buoA | 0.17 | 0.20 | 0.92 | 2.70 | Download | ----NSD---------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPIV------DIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTYHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL----- | |||||||||||||||||||
| 9 | 5tgzA | 0.20 | 0.23 | 0.90 | 3.33 | Download | -GGRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG-------NCEKLQSV-----------CSDIHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 10 | 4gpoA | 0.19 | 0.20 | 0.88 | 4.86 | Download | ---------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIIFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL------ | |||||||||||||||||||
| ||||||||||||||||||||||||||
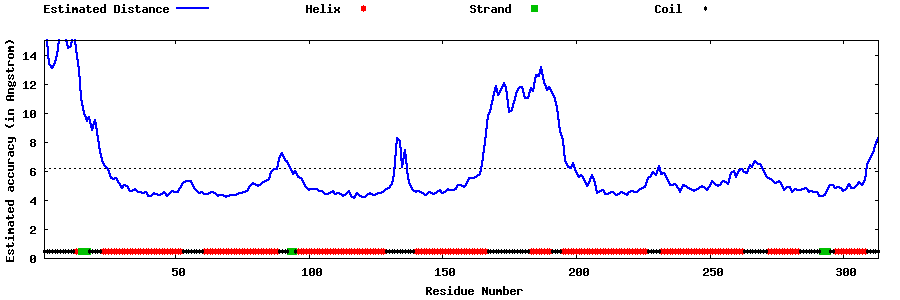
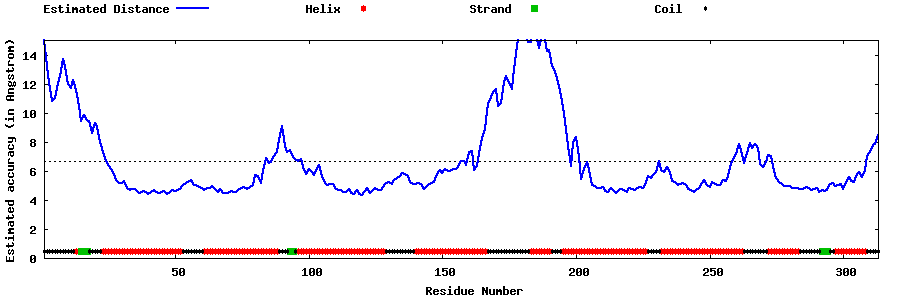
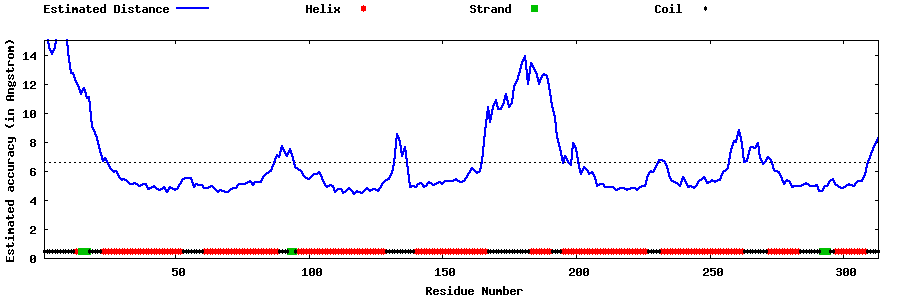
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||